Diana Mateus
LS2N - équipe SIMS, LS2N, CFE, Nantes Univ - ECN, Nantes Univ
Implicit Shape-Prior for Few-Shot Assisted 3D Segmentation
Sep 10, 2025Abstract:The objective of this paper is to significantly reduce the manual workload required from medical professionals in complex 3D segmentation tasks that cannot be yet fully automated. For instance, in radiotherapy planning, organs at risk must be accurately identified in computed tomography (CT) or magnetic resonance imaging (MRI) scans to ensure they are spared from harmful radiation. Similarly, diagnosing age-related degenerative diseases such as sarcopenia, which involve progressive muscle volume loss and strength, is commonly based on muscular mass measurements often obtained from manual segmentation of medical volumes. To alleviate the manual-segmentation burden, this paper introduces an implicit shape prior to segment volumes from sparse slice manual annotations generalized to the multi-organ case, along with a simple framework for automatically selecting the most informative slices to guide and minimize the next interactions. The experimental validation shows the method's effectiveness on two medical use cases: assisted segmentation in the context of at risks organs for brain cancer patients, and acceleration of the creation of a new database with unseen muscle shapes for patients with sarcopenia.
Unsupervised Anomaly Detection on Implicit Shape representations for Sarcopenia Detection
Feb 13, 2025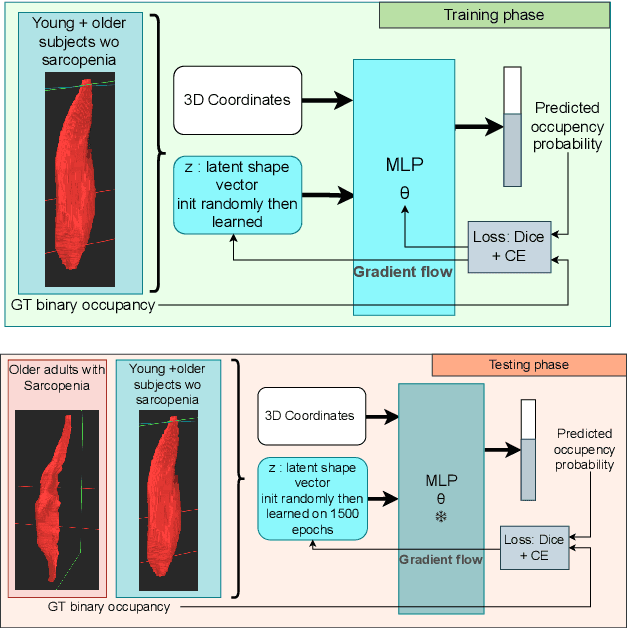
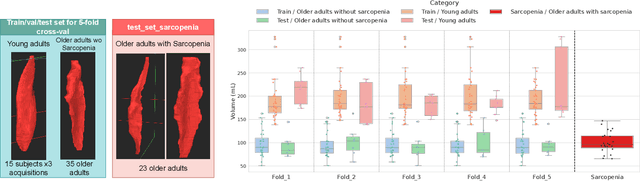
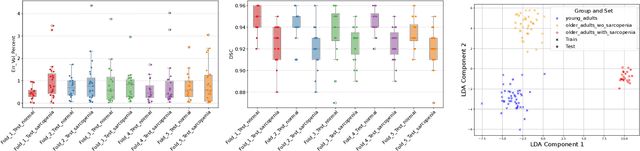
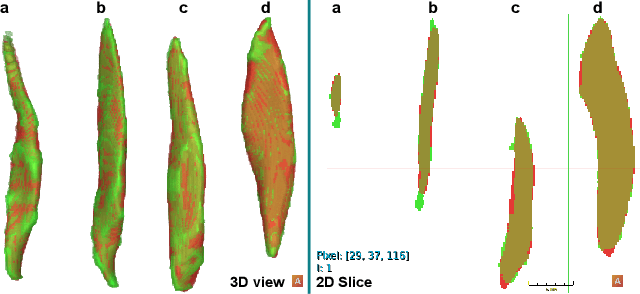
Abstract:Sarcopenia is an age-related progressive loss of muscle mass and strength that significantly impacts daily life. A commonly studied criterion for characterizing the muscle mass has been the combination of 3D imaging and manual segmentations. In this paper, we instead study the muscles' shape. We rely on an implicit neural representation (INR) to model normal muscle shapes. We then introduce an unsupervised anomaly detection method to identify sarcopenic muscles based on the reconstruction error of the implicit model. Relying on a conditional INR with an auto-decoding strategy, we also learn a latent representation of the muscles that clearly separates normal from abnormal muscles in an unsupervised fashion. Experimental results on a dataset of 103 segmented volumes indicate that our double anomaly detection strategy effectively discriminates sarcopenic and non-sarcopenic muscles.
Ultrasound Image Enhancement with the Variance of Diffusion Models
Sep 17, 2024



Abstract:Ultrasound imaging, despite its widespread use in medicine, often suffers from various sources of noise and artifacts that impact the signal-to-noise ratio and overall image quality. Enhancing ultrasound images requires a delicate balance between contrast, resolution, and speckle preservation. This paper introduces a novel approach that integrates adaptive beamforming with denoising diffusion-based variance imaging to address this challenge. By applying Eigenspace-Based Minimum Variance (EBMV) beamforming and employing a denoising diffusion model fine-tuned on ultrasound data, our method computes the variance across multiple diffusion-denoised samples to produce high-quality despeckled images. This approach leverages both the inherent multiplicative noise of ultrasound and the stochastic nature of diffusion models. Experimental results on a publicly available dataset demonstrate the effectiveness of our method in achieving superior image reconstructions from single plane-wave acquisitions. The code is available at: https://github.com/Yuxin-Zhang-Jasmine/IUS2024_Diffusion.
Compact Implicit Neural Representations for Plane Wave Images
Sep 17, 2024



Abstract:Ultrafast Plane-Wave (PW) imaging often produces artifacts and shadows that vary with insonification angles. We propose a novel approach using Implicit Neural Representations (INRs) to compactly encode multi-planar sequences while preserving crucial orientation-dependent information. To our knowledge, this is the first application of INRs for PW angular interpolation. Our method employs a Multi-Layer Perceptron (MLP)-based model with a concise physics-enhanced rendering technique. Quantitative evaluations using SSIM, PSNR, and standard ultrasound metrics, along with qualitative visual assessments, confirm the effectiveness of our approach. Additionally, our method demonstrates significant storage efficiency, with model weights requiring 530 KB compared to 8 MB for directly storing the 75 PW images, achieving a notable compression ratio of approximately 15:1.
Ultrasound Imaging based on the Variance of a Diffusion Restoration Model
Mar 22, 2024



Abstract:Despite today's prevalence of ultrasound imaging in medicine, ultrasound signal-to-noise ratio is still affected by several sources of noise and artefacts. Moreover, enhancing ultrasound image quality involves balancing concurrent factors like contrast, resolution, and speckle preservation. Recently, there has been progress in both model-based and learning-based approaches addressing the problem of ultrasound image reconstruction. Bringing the best from both worlds, we propose a hybrid reconstruction method combining an ultrasound linear direct model with a learning-based prior coming from a generative Denoising Diffusion model. More specifically, we rely on the unsupervised fine-tuning of a pre-trained Denoising Diffusion Restoration Model (DDRM). Given the nature of multiplicative noise inherent to ultrasound, this paper proposes an empirical model to characterize the stochasticity of diffusion reconstruction of ultrasound images, and shows the interest of its variance as an echogenicity map estimator. We conduct experiments on synthetic, in-vitro, and in-vivo data, demonstrating the efficacy of our variance imaging approach in achieving high-quality image reconstructions from single plane-wave acquisitions and in comparison to state-of-the-art methods.
Muscle volume quantification: guiding transformers with anatomical priors
Oct 31, 2023Abstract:Muscle volume is a useful quantitative biomarker in sports, but also for the follow-up of degenerative musculo-skelletal diseases. In addition to volume, other shape biomarkers can be extracted by segmenting the muscles of interest from medical images. Manual segmentation is still today the gold standard for such measurements despite being very time-consuming. We propose a method for automatic segmentation of 18 muscles of the lower limb on 3D Magnetic Resonance Images to assist such morphometric analysis. By their nature, the tissue of different muscles is undistinguishable when observed in MR Images. Thus, muscle segmentation algorithms cannot rely on appearance but only on contour cues. However, such contours are hard to detect and their thickness varies across subjects. To cope with the above challenges, we propose a segmentation approach based on a hybrid architecture, combining convolutional and visual transformer blocks. We investigate for the first time the behaviour of such hybrid architectures in the context of muscle segmentation for shape analysis. Considering the consistent anatomical muscle configuration, we rely on transformer blocks to capture the longrange relations between the muscles. To further exploit the anatomical priors, a second contribution of this work consists in adding a regularisation loss based on an adjacency matrix of plausible muscle neighbourhoods estimated from the training data. Our experimental results on a unique database of elite athletes show it is possible to train complex hybrid models from a relatively small database of large volumes, while the anatomical prior regularisation favours better predictions.
Diffusion Reconstruction of Ultrasound Images with Informative Uncertainty
Oct 31, 2023



Abstract:Despite its wide use in medicine, ultrasound imaging faces several challenges related to its poor signal-to-noise ratio and several sources of noise and artefacts. Enhancing ultrasound image quality involves balancing concurrent factors like contrast, resolution, and speckle preservation. In recent years, there has been progress both in model-based and learning-based approaches to improve ultrasound image reconstruction. Bringing the best from both worlds, we propose a hybrid approach leveraging advances in diffusion models. To this end, we adapt Denoising Diffusion Restoration Models (DDRM) to incorporate ultrasound physics through a linear direct model and an unsupervised fine-tuning of the prior diffusion model. We conduct comprehensive experiments on simulated, in-vitro, and in-vivo data, demonstrating the efficacy of our approach in achieving high-quality image reconstructions from a single plane wave input and in comparison to state-of-the-art methods. Finally, given the stochastic nature of the method, we analyse in depth the statistical properties of single and multiple-sample reconstructions, experimentally show the informativeness of their variance, and provide an empirical model relating this behaviour to speckle noise. The code and data are available at: (upon acceptance).
Graph-based multimodal multi-lesion DLBCL treatment response prediction from PET images
Oct 25, 2023Abstract:Diffuse Large B-cell Lymphoma (DLBCL) is a lymphatic cancer involving one or more lymph nodes and extranodal sites. Its diagnostic and follow-up rely on Positron Emission Tomography (PET) and Computed Tomography (CT). After diagnosis, the number of nonresponding patients to standard front-line therapy remains significant (30-40%). This work aims to develop a computer-aided approach to identify high-risk patients requiring adapted treatment by efficiently exploiting all the information available for each patient, including both clinical and image data. We propose a method based on recent graph neural networks that combine imaging information from multiple lesions, and a cross-attention module to integrate different data modalities efficiently. The model is trained and evaluated on a private prospective multicentric dataset of 583 patients. Experimental results show that our proposed method outperforms classical supervised methods based on either clinical, imaging or both clinical and imaging data for the 2-year progression-free survival (PFS) classification accuracy.
Can ultrasound confidence maps predict sonographers' labeling variability?
Aug 18, 2023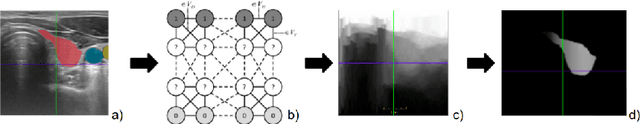
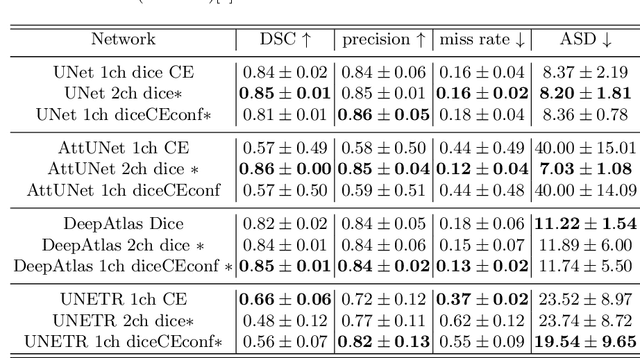
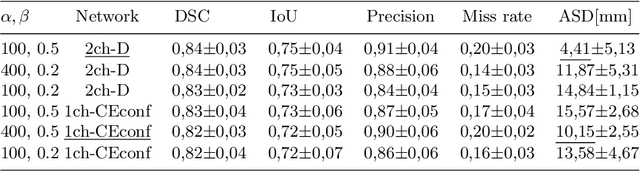
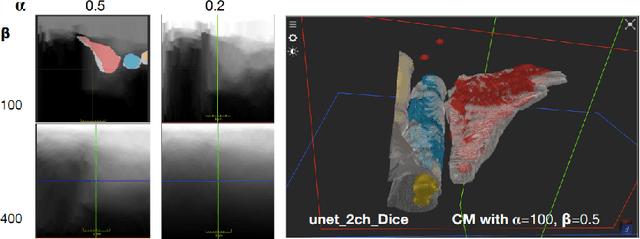
Abstract:Measuring cross-sectional areas in ultrasound images is a standard tool to evaluate disease progress or treatment response. Often addressed today with supervised deep-learning segmentation approaches, existing solutions highly depend upon the quality of experts' annotations. However, the annotation quality in ultrasound is anisotropic and position-variant due to the inherent physical imaging principles, including attenuation, shadows, and missing boundaries, commonly exacerbated with depth. This work proposes a novel approach that guides ultrasound segmentation networks to account for sonographers' uncertainties and generate predictions with variability similar to the experts. We claim that realistic variability can reduce overconfident predictions and improve physicians' acceptance of deep-learning cross-sectional segmentation solutions. Our method provides CM's certainty for each pixel for minimal computational overhead as it can be precalculated directly from the image. We show that there is a correlation between low values in the confidence maps and expert's label uncertainty. Therefore, we propose to give the confidence maps as additional information to the networks. We study the effect of the proposed use of ultrasound CMs in combination with four state-of-the-art neural networks and in two configurations: as a second input channel and as part of the loss. We evaluate our method on 3D ultrasound datasets of the thyroid and lower limb muscles. Our results show ultrasound CMs increase the Dice score, improve the Hausdorff and Average Surface Distances, and decrease the number of isolated pixel predictions. Furthermore, our findings suggest that ultrasound CMs improve the penalization of uncertain areas in the ground truth data, thereby improving problematic interpolations. Our code and example data will be made public at https://github.com/IFL-CAMP/Confidence-segmentation.
Ultrasound Image Reconstruction with Denoising Diffusion Restoration Models
Jul 29, 2023



Abstract:Ultrasound image reconstruction can be approximately cast as a linear inverse problem that has traditionally been solved with penalized optimization using the $l_1$ or $l_2$ norm, or wavelet-based terms. However, such regularization functions often struggle to balance the sparsity and the smoothness. A promising alternative is using learned priors to make the prior knowledge closer to reality. In this paper, we rely on learned priors under the framework of Denoising Diffusion Restoration Models (DDRM), initially conceived for restoration tasks with natural images. We propose and test two adaptions of DDRM to ultrasound inverse problem models, DRUS and WDRUS. Our experiments on synthetic and PICMUS data show that from a single plane wave our method can achieve image quality comparable to or better than DAS and state-of-the-art methods. The code is available at: https://github.com/Yuxin-Zhang-Jasmine/DRUS-v1.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge