Gianmarco Santini
Deformable Image Registration with Deep Network Priors: a Study on Longitudinal PET Images
Nov 22, 2021



Abstract:Longitudinal image registration is challenging and has not yet benefited from major performance improvements thanks to deep-learning. Inspired by Deep Image Prior, this paper introduces a different use of deep architectures as regularizers to tackle the image registration question. We propose a subject-specific deformable registration method called MIRRBA, relying on a deep pyramidal architecture to be the prior parametric model constraining the deformation field. Diverging from the supervised learning paradigm, MIRRBA does not require a learning database, but only the pair of images to be registered to optimize the network's parameters and provide a deformation field. We demonstrate the regularizing power of deep architectures and present new elements to understand the role of the architecture in deep learning methods for registration. Hence, to study the impact of the network parameters, we ran our method with different architectural configurations on a private dataset of 110 metastatic breast cancer full-body PET images with manual segmentations of the brain, bladder and metastatic lesions. We compared it against conventional iterative registration approaches and supervised deep learning-based models. Global and local registration accuracies were evaluated using the detection rate and the Dice score respectively, while registration realism was evaluated using the Jacobian's determinant. Moreover, we computed the ability of the different methods to shrink vanishing lesions with the disappearing rate. MIRRBA significantly improves the organ and lesion Dice scores of supervised models. Regarding the disappearing rate, MIRRBA more than doubles the best performing conventional approach SyNCC score. Our work therefore proposes an alternative way to bridge the performance gap between conventional and deep learning-based methods and demonstrates the regularizing power of deep architectures.
Kidney tumor segmentation using an ensembling multi-stage deep learning approach. A contribution to the KiTS19 challenge
Sep 02, 2019



Abstract:Precise characterization of the kidney and kidney tumor characteristics is of outmost importance in the context of kidney cancer treatment, especially for nephron sparing surgery which requires a precise localization of the tissues to be removed. The need for accurate and automatic delineation tools is at the origin of the KiTS19 challenge. It aims at accelerating the research and development in this field to aid prognosis and treatment planning by providing a characterized dataset of 300 CT scans to be segmented. To address the challenge, we proposed an automatic, multi-stage, 2.5D deep learning-based segmentation approach based on Residual UNet framework. An ensembling operation is added at the end to combine prediction results from previous stages reducing the variance between single models. Our neural network segmentation algorithm reaches a mean Dice score of 0.96 and 0.74 for kidney and kidney tumors, respectively on 90 unseen test cases. The results obtained are promising and could be improved by incorporating prior knowledge about the benign cysts that regularly lower the tumor segmentation results.
Unsupervised Data Selection for Supervised Learning
Oct 29, 2018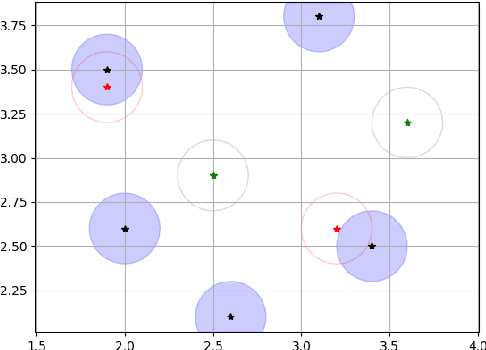
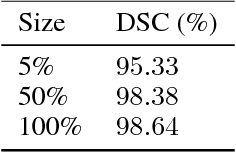
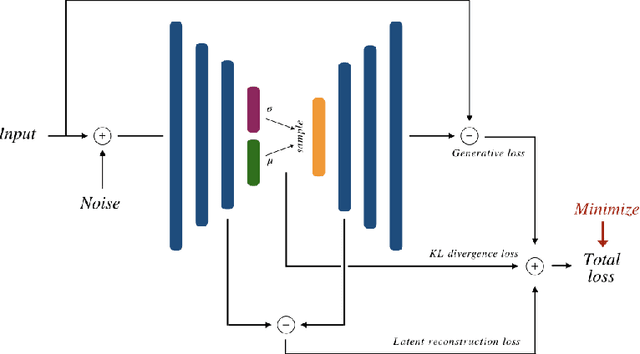
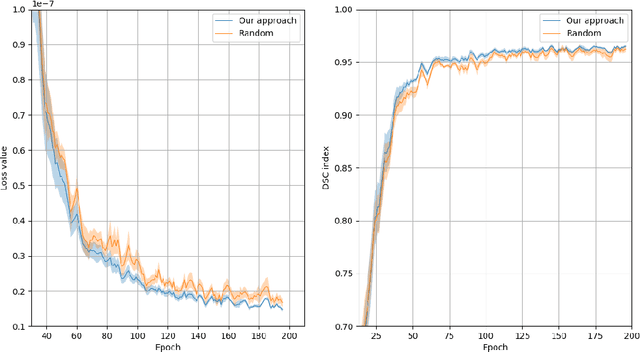
Abstract:Recent research put a big effort in the development of deep learning architectures and optimizers obtaining impressive results in areas ranging from vision to language processing. However little attention has been addressed to the need of a methodological process of data collection. In this work we show that high quality data for supervised learning can be selected in an unsupervised manner and that by doing so one can obtain models capable to generalize better than in the case of random training set construction.
Training of a Skull-Stripping Neural Network with efficient data augmentation
Oct 25, 2018
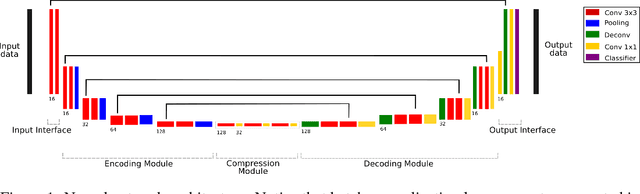

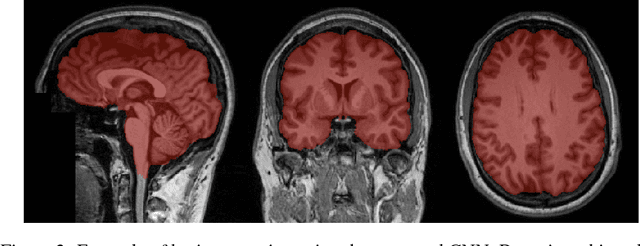
Abstract:Skull-stripping methods aim to remove the non-brain tissue from acquisition of brain scans in magnetic resonance (MR) imaging. Although several methods sharing this common purpose have been presented in literature, they all suffer from the great variability of the MR images. In this work we propose a novel approach based on Convolutional Neural Networks to automatically perform the brain extraction obtaining cutting-edge performance in the NFBS public database. Additionally, we focus on the efficient training of the neural network designing an effective data augmentation pipeline. Obtained results are evaluated through Dice metric, obtaining a value of 96.5%, and processing time, with 4.5s per volume.
Convolutional Neural Networks for the segmentation of microcalcification in Mammography Imaging
Sep 11, 2018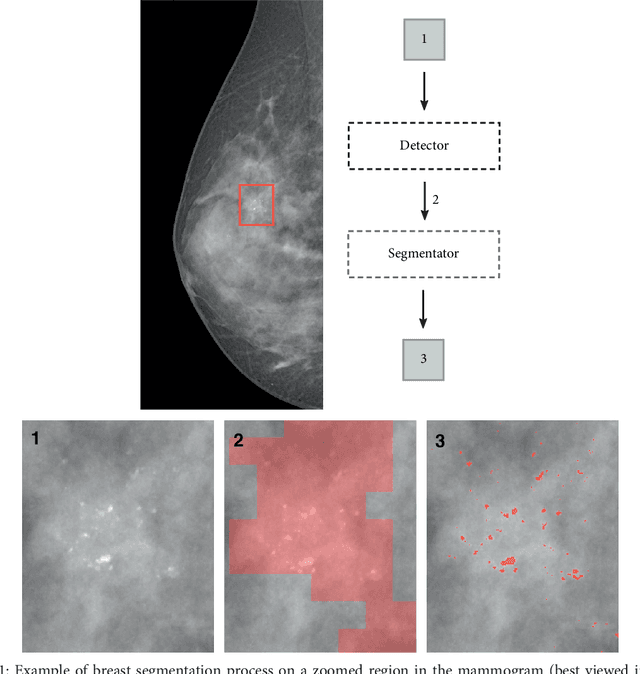

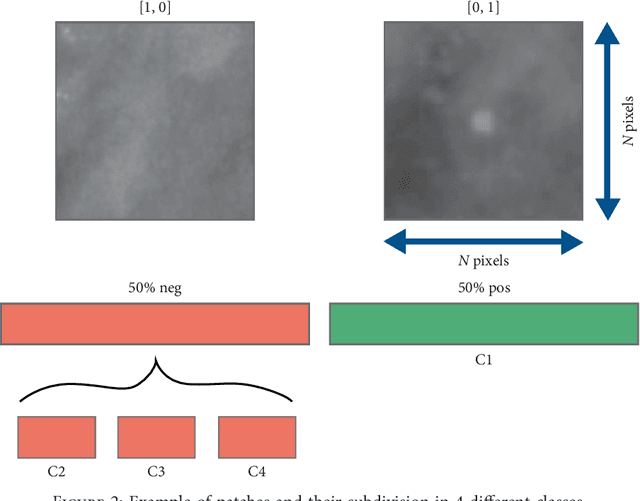
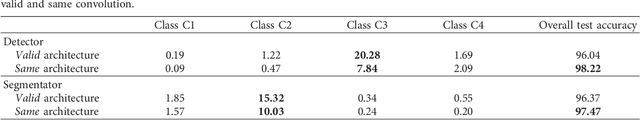
Abstract:Cluster of microcalcifications can be an early sign of breast cancer. In this paper we propose a novel approach based on convolutional neural networks for the detection and segmentation of microcalcification clusters. In this work we used 283 mammograms to train and validate our model, obtaining an accuracy of 98.22% in the detection of preliminary suspect regions and of 97.47% in the segmentation task. Our results show how deep learning could be an effective tool to effectively support radiologists during mammograms examination.
Synthetic contrast enhancement in cardiac CT with Deep Learning
Jul 02, 2018



Abstract:In Europe the 20% of the CT scans cover the thoracic region. The acquired images contain information about the cardiovascular system that often remains latent due to the lack of contrast in the cardiac area. On the other hand, the contrast enhanced computed tomography (CECT) represents an imaging technique that allows to easily assess the cardiac chambers volumes and the contrast dynamics. With this work we aim to face the problem of extraction and presentation of these latent information, using a deep learning approach with convolutional neural networks. Starting from the extraction of relevant features from the image without contrast medium, we try to re-map them on features typical of CECT, to synthesize an image characterized by an attenuation in the cardiac chambers as if a virtually iodine contrast medium was injected. The purposes are to guarantee an estimation of the left cardiac chambers volume and to perform an evaluation of the contrast dynamics. Our approach is based on a deconvolutional network trained on a set of 120 patients who underwent both CT acquisitions in the same contrastographic arterial phase and the same cardiac phase. To ensure a reliable predicted CECT image, in terms of values and morphology, a custom loss function is defined by combining an error function to find a pixel-wise correspondence, which takes into account the similarity in term of Hounsfield units between the input and output images and by a cross-entropy computed on the binarized versions of the synthesized and of the real CECT image. The proposed method is finally tested on 20 subjects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge