Andrea Leo
Re-using Adversarial Mask Discriminators for Test-time Training under Distribution Shifts
Aug 26, 2021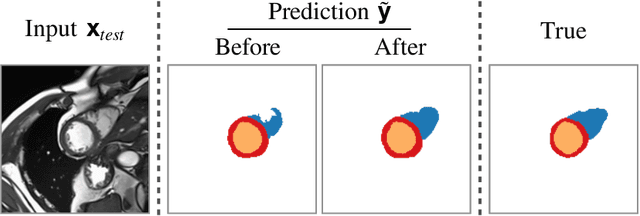
Abstract:Thanks to their ability to learn flexible data-driven losses, Generative Adversarial Networks (GANs) are an integral part of many semi- and weakly-supervised methods for medical image segmentation. GANs jointly optimise a generator and an adversarial discriminator on a set of training data. After training has completed, the discriminator is usually discarded and only the generator is used for inference. But should we discard discriminators? In this work, we argue that training stable discriminators produces expressive loss functions that we can re-use at inference to detect and correct segmentation mistakes. First, we identify key challenges and suggest possible solutions to make discriminators re-usable at inference. Then, we show that we can combine discriminators with image reconstruction costs (via decoders) to further improve the model. Our method is simple and improves the test-time performance of pre-trained GANs. Moreover, we show that it is compatible with standard post-processing techniques and it has potentials to be used for Online Continual Learning. With our work, we open new research avenues for re-using adversarial discriminators at inference.
Stop Throwing Away Discriminators! Re-using Adversaries for Test-Time Training
Aug 26, 2021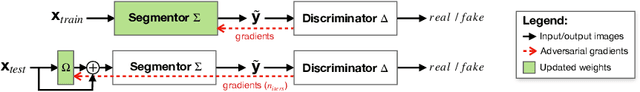
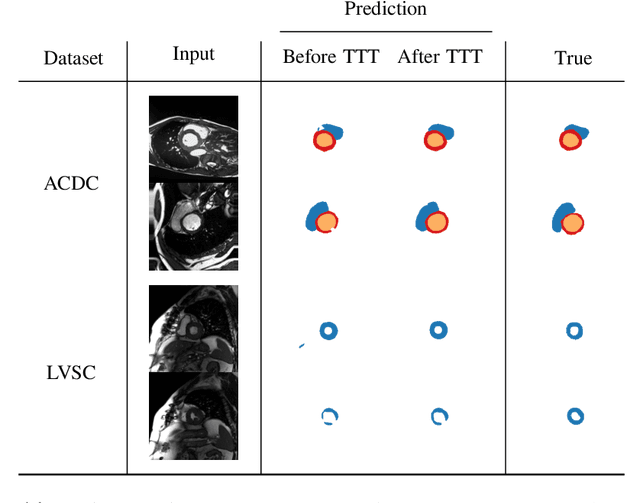
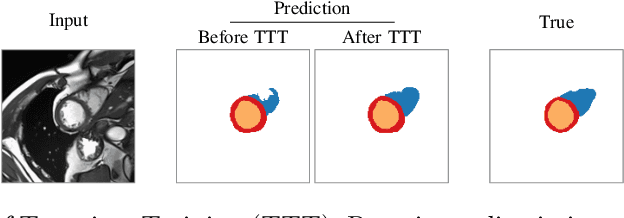

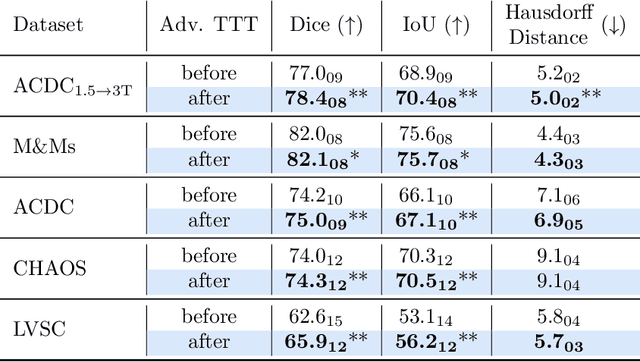
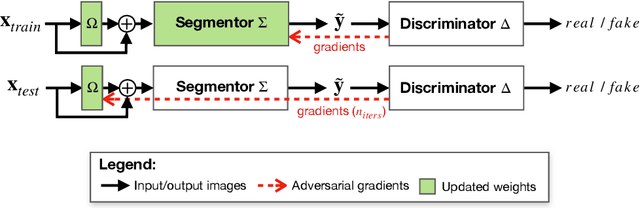
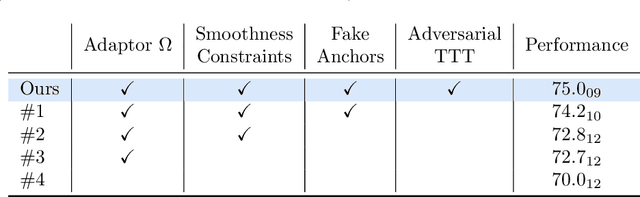
Abstract:Thanks to their ability to learn data distributions without requiring paired data, Generative Adversarial Networks (GANs) have become an integral part of many computer vision methods, including those developed for medical image segmentation. These methods jointly train a segmentor and an adversarial mask discriminator, which provides a data-driven shape prior. At inference, the discriminator is discarded, and only the segmentor is used to predict label maps on test images. But should we discard the discriminator? Here, we argue that the life cycle of adversarial discriminators should not end after training. On the contrary, training stable GANs produces powerful shape priors that we can use to correct segmentor mistakes at inference. To achieve this, we develop stable mask discriminators that do not overfit or catastrophically forget. At test time, we fine-tune the segmentor on each individual test instance until it satisfies the learned shape prior. Our method is simple to implement and increases model performance. Moreover, it opens new directions for re-using mask discriminators at inference. We release the code used for the experiments at https://vios-s.github.io/adversarial-test-time-training.
Self-supervised Multi-scale Consistency for Weakly Supervised Segmentation Learning
Aug 26, 2021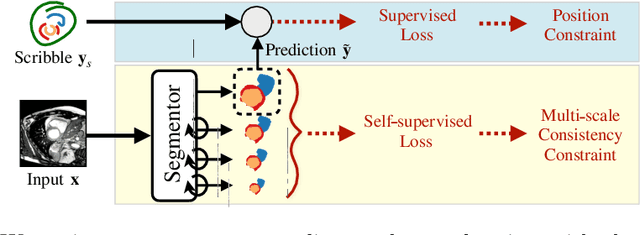
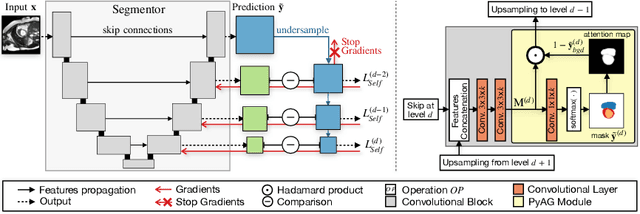
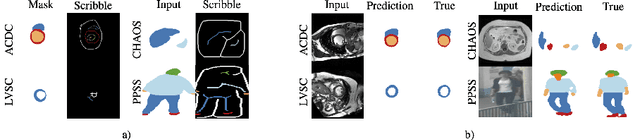
Abstract:Collecting large-scale medical datasets with fine-grained annotations is time-consuming and requires experts. For this reason, weakly supervised learning aims at optimising machine learning models using weaker forms of annotations, such as scribbles, which are easier and faster to collect. Unfortunately, training with weak labels is challenging and needs regularisation. Herein, we introduce a novel self-supervised multi-scale consistency loss, which, coupled with an attention mechanism, encourages the segmentor to learn multi-scale relationships between objects and improves performance. We show state-of-the-art performance on several medical and non-medical datasets. The code used for the experiments is available at https://vios-s.github.io/multiscale-pyag.
Weakly Supervised Segmentation with Multi-scale Adversarial Attention Gates
Jul 02, 2020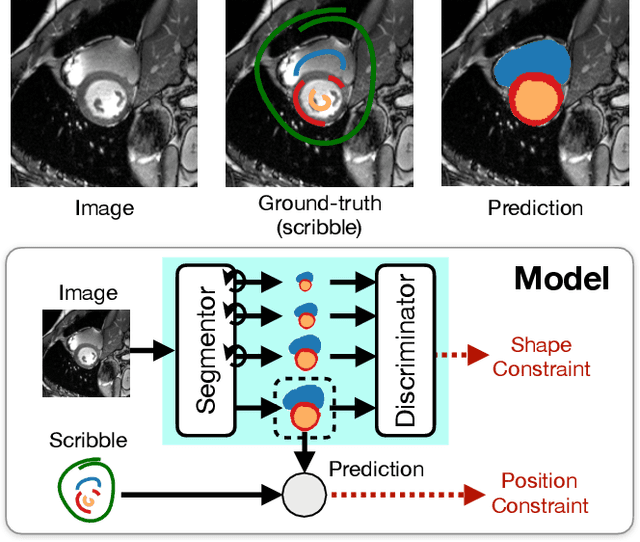

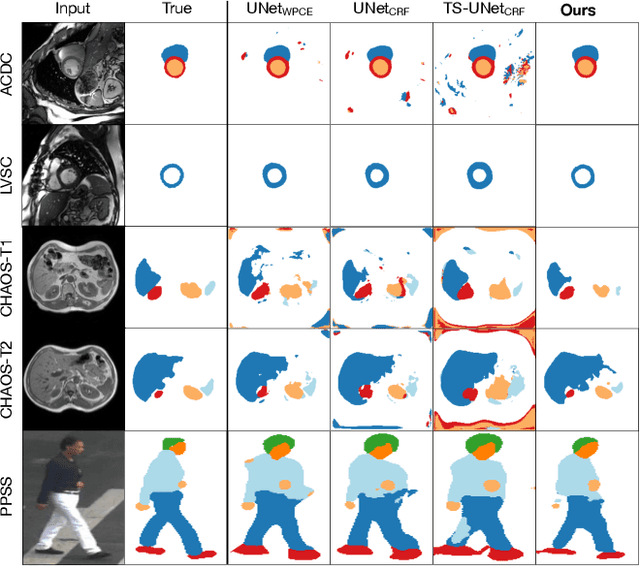
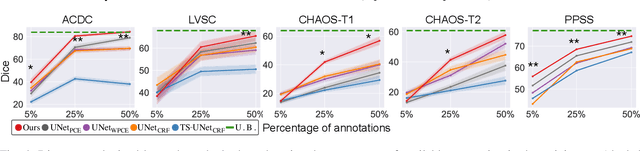
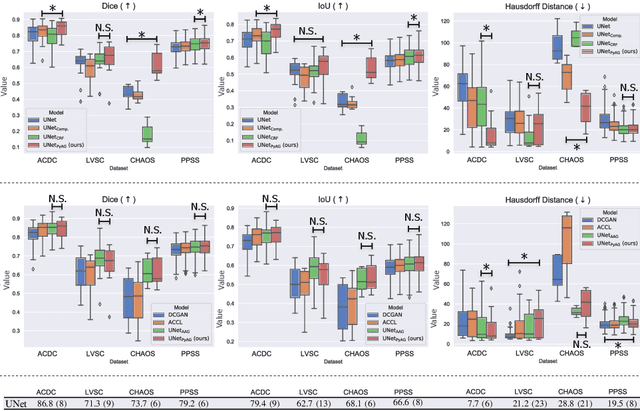
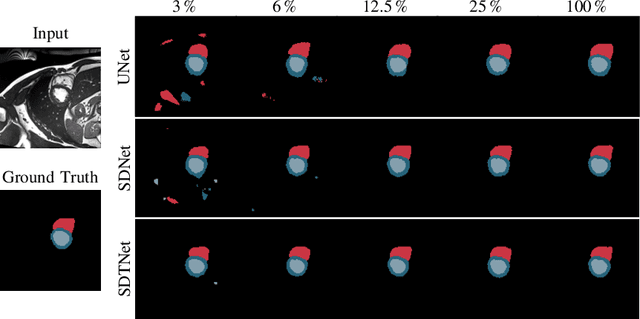
Abstract:Large, fine-grained image segmentation datasets, annotated at pixel-level, are difficult to obtain, particularly in medical imaging, where annotations also require expert knowledge. Weakly-supervised learning can train models by relying on weaker forms of annotation, such as scribbles. Here, we learn to segment using scribble annotations in an adversarial game. With unpaired segmentation masks, we train a multi-scale GAN to generate realistic segmentation masks at multiple resolutions, while we use scribbles to learn the correct position in the image. Central to the model's success is a novel attention gating mechanism, which we condition with adversarial signals to act as a shape prior, resulting in better object localization at multiple scales. We evaluated our model on several medical (ACDC, LVSC, CHAOS) and non-medical (PPSS) datasets, and we report performance levels matching those achieved by models trained with fully annotated segmentation masks. We also demonstrate extensions in a variety of settings: semi-supervised learning; combining multiple scribble sources (a crowdsourcing scenario) and multi-task learning (combining scribble and mask supervision). We will release expert-made scribble annotations for the ACDC dataset, and the code used for the experiments, at https://gvalvano.github.io/wss-multiscale-adversarial-attention-gates.
Temporal Consistency Objectives Regularize the Learning of Disentangled Representations
Aug 29, 2019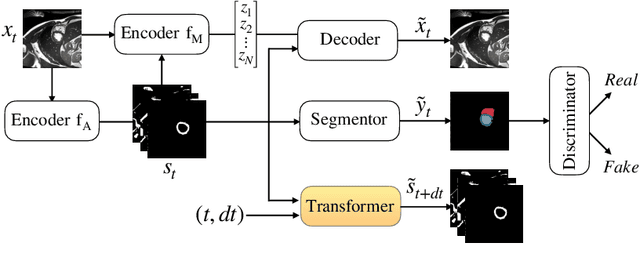
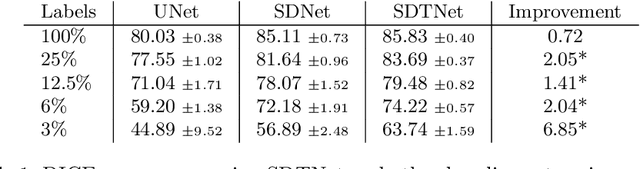

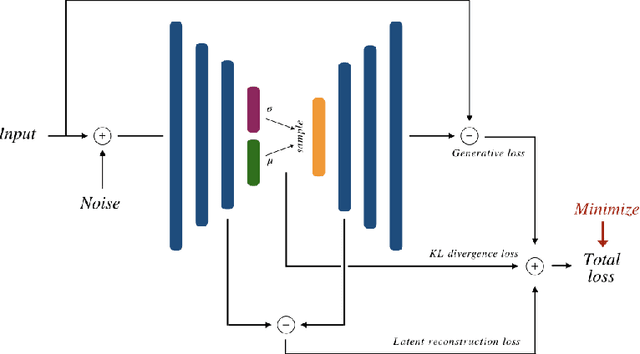
Abstract:There has been an increasing focus in learning interpretable feature representations, particularly in applications such as medical image analysis that require explainability, whilst relying less on annotated data (since annotations can be tedious and costly). Here we build on recent innovations in style-content representations to learn anatomy, imaging characteristics (appearance) and temporal correlations. By introducing a self-supervised objective of predicting future cardiac phases we improve disentanglement. We propose a temporal transformer architecture that given an image conditioned on phase difference, it predicts a future frame. This forces the anatomical decomposition to be consistent with the temporal cardiac contraction in cine MRI and to have semantic meaning with less need for annotations. We demonstrate that using this regularization, we achieve competitive results and improve semi-supervised segmentation, especially when very few labelled data are available. Specifically, we show Dice increase of up to 19\% and 7\% compared to supervised and semi-supervised approaches respectively on the ACDC dataset. Code is available at: https://github.com/gvalvano/sdtnet .
Unsupervised Data Selection for Supervised Learning
Oct 29, 2018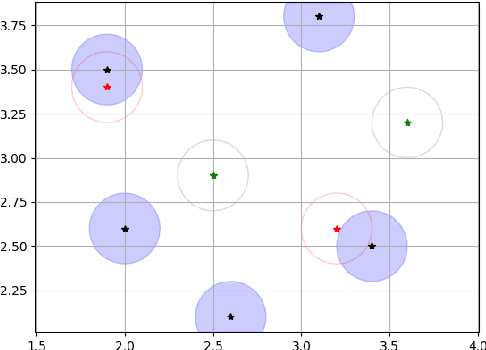
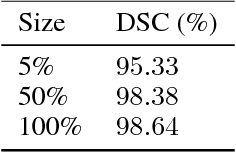
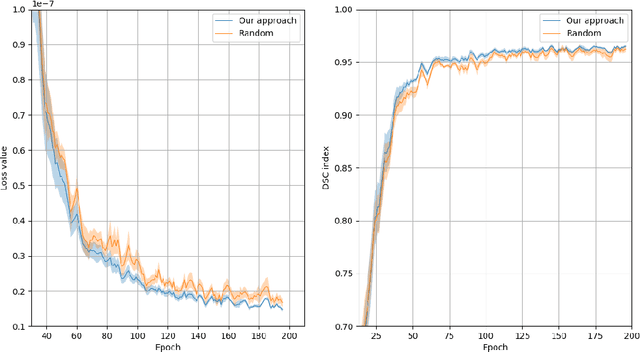
Abstract:Recent research put a big effort in the development of deep learning architectures and optimizers obtaining impressive results in areas ranging from vision to language processing. However little attention has been addressed to the need of a methodological process of data collection. In this work we show that high quality data for supervised learning can be selected in an unsupervised manner and that by doing so one can obtain models capable to generalize better than in the case of random training set construction.
Training of a Skull-Stripping Neural Network with efficient data augmentation
Oct 25, 2018
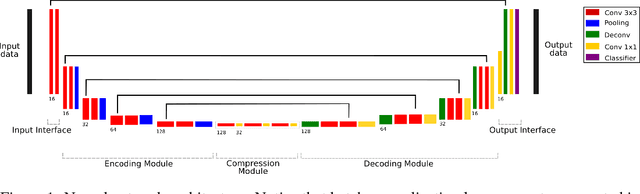

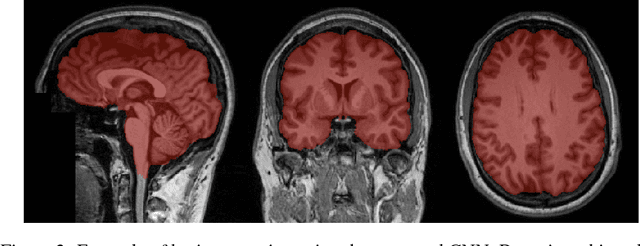
Abstract:Skull-stripping methods aim to remove the non-brain tissue from acquisition of brain scans in magnetic resonance (MR) imaging. Although several methods sharing this common purpose have been presented in literature, they all suffer from the great variability of the MR images. In this work we propose a novel approach based on Convolutional Neural Networks to automatically perform the brain extraction obtaining cutting-edge performance in the NFBS public database. Additionally, we focus on the efficient training of the neural network designing an effective data augmentation pipeline. Obtained results are evaluated through Dice metric, obtaining a value of 96.5%, and processing time, with 4.5s per volume.
Synthetic contrast enhancement in cardiac CT with Deep Learning
Jul 02, 2018



Abstract:In Europe the 20% of the CT scans cover the thoracic region. The acquired images contain information about the cardiovascular system that often remains latent due to the lack of contrast in the cardiac area. On the other hand, the contrast enhanced computed tomography (CECT) represents an imaging technique that allows to easily assess the cardiac chambers volumes and the contrast dynamics. With this work we aim to face the problem of extraction and presentation of these latent information, using a deep learning approach with convolutional neural networks. Starting from the extraction of relevant features from the image without contrast medium, we try to re-map them on features typical of CECT, to synthesize an image characterized by an attenuation in the cardiac chambers as if a virtually iodine contrast medium was injected. The purposes are to guarantee an estimation of the left cardiac chambers volume and to perform an evaluation of the contrast dynamics. Our approach is based on a deconvolutional network trained on a set of 120 patients who underwent both CT acquisitions in the same contrastographic arterial phase and the same cardiac phase. To ensure a reliable predicted CECT image, in terms of values and morphology, a custom loss function is defined by combining an error function to find a pixel-wise correspondence, which takes into account the similarity in term of Hounsfield units between the input and output images and by a cross-entropy computed on the binarized versions of the synthesized and of the real CECT image. The proposed method is finally tested on 20 subjects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge