Noemie Moreau
KPIs 2024 Challenge: Advancing Glomerular Segmentation from Patch- to Slide-Level
Feb 11, 2025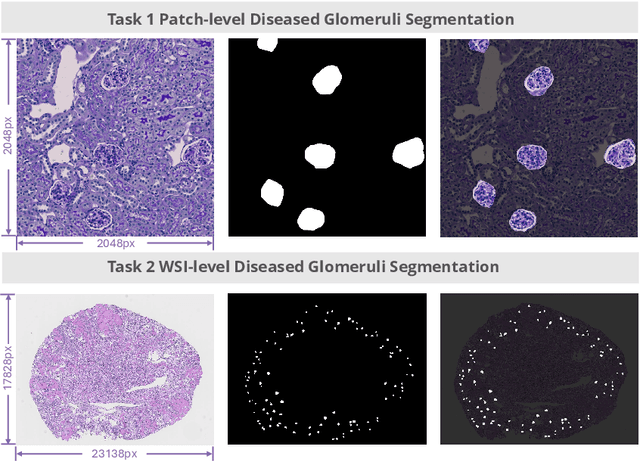
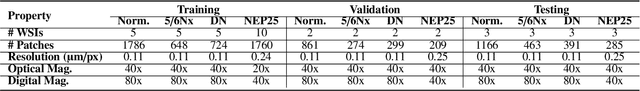
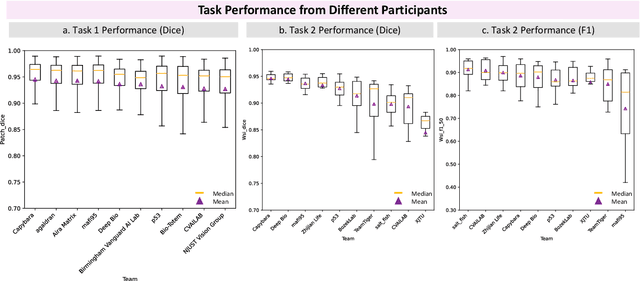

Abstract:Chronic kidney disease (CKD) is a major global health issue, affecting over 10% of the population and causing significant mortality. While kidney biopsy remains the gold standard for CKD diagnosis and treatment, the lack of comprehensive benchmarks for kidney pathology segmentation hinders progress in the field. To address this, we organized the Kidney Pathology Image Segmentation (KPIs) Challenge, introducing a dataset that incorporates preclinical rodent models of CKD with over 10,000 annotated glomeruli from 60+ Periodic Acid Schiff (PAS)-stained whole slide images. The challenge includes two tasks, patch-level segmentation and whole slide image segmentation and detection, evaluated using the Dice Similarity Coefficient (DSC) and F1-score. By encouraging innovative segmentation methods that adapt to diverse CKD models and tissue conditions, the KPIs Challenge aims to advance kidney pathology analysis, establish new benchmarks, and enable precise, large-scale quantification for disease research and diagnosis.
Deformable Image Registration with Deep Network Priors: a Study on Longitudinal PET Images
Nov 22, 2021



Abstract:Longitudinal image registration is challenging and has not yet benefited from major performance improvements thanks to deep-learning. Inspired by Deep Image Prior, this paper introduces a different use of deep architectures as regularizers to tackle the image registration question. We propose a subject-specific deformable registration method called MIRRBA, relying on a deep pyramidal architecture to be the prior parametric model constraining the deformation field. Diverging from the supervised learning paradigm, MIRRBA does not require a learning database, but only the pair of images to be registered to optimize the network's parameters and provide a deformation field. We demonstrate the regularizing power of deep architectures and present new elements to understand the role of the architecture in deep learning methods for registration. Hence, to study the impact of the network parameters, we ran our method with different architectural configurations on a private dataset of 110 metastatic breast cancer full-body PET images with manual segmentations of the brain, bladder and metastatic lesions. We compared it against conventional iterative registration approaches and supervised deep learning-based models. Global and local registration accuracies were evaluated using the detection rate and the Dice score respectively, while registration realism was evaluated using the Jacobian's determinant. Moreover, we computed the ability of the different methods to shrink vanishing lesions with the disappearing rate. MIRRBA significantly improves the organ and lesion Dice scores of supervised models. Regarding the disappearing rate, MIRRBA more than doubles the best performing conventional approach SyNCC score. Our work therefore proposes an alternative way to bridge the performance gap between conventional and deep learning-based methods and demonstrates the regularizing power of deep architectures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge