Mathieu Rubeaux
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021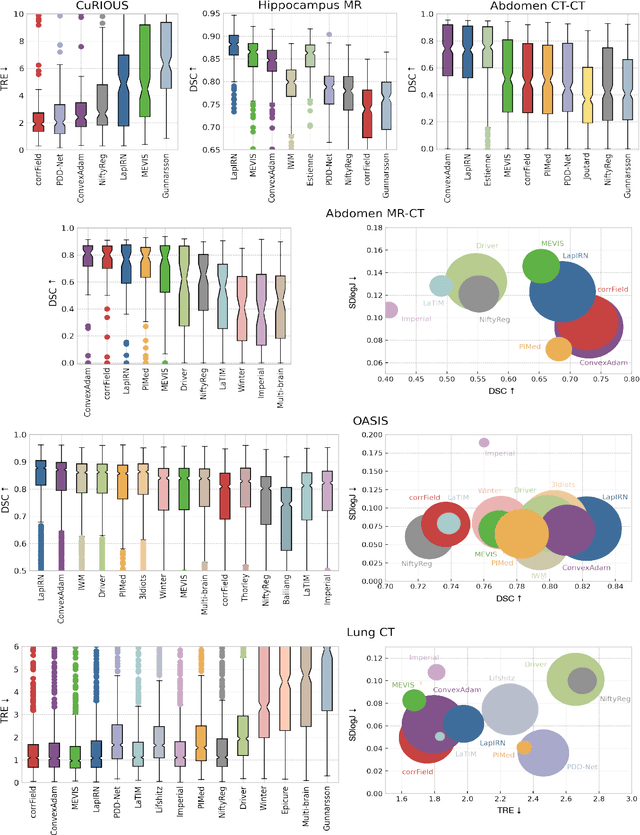
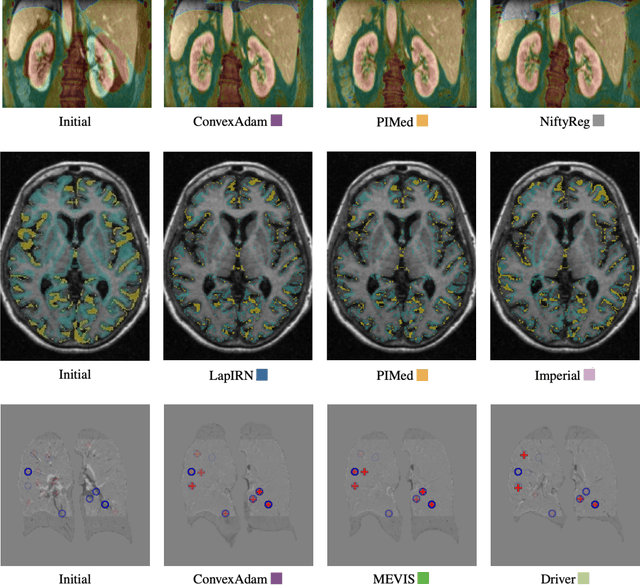
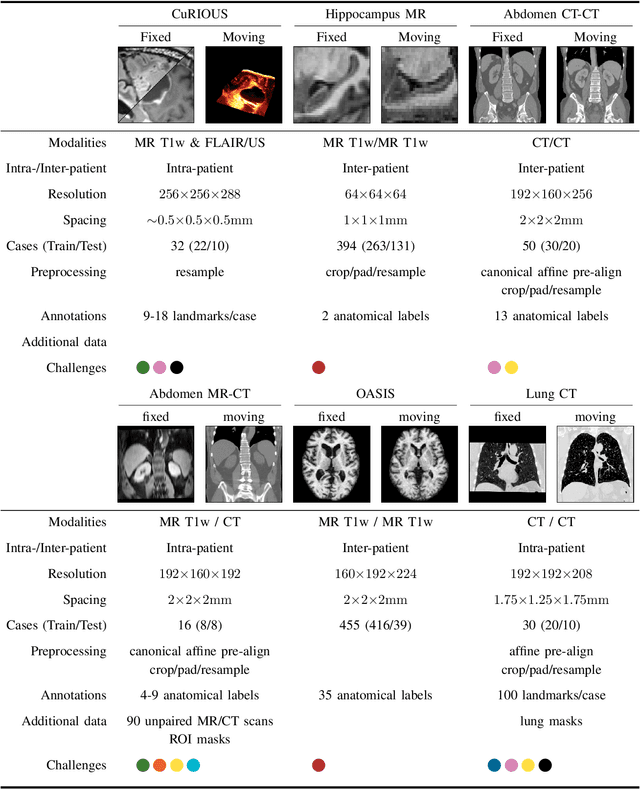
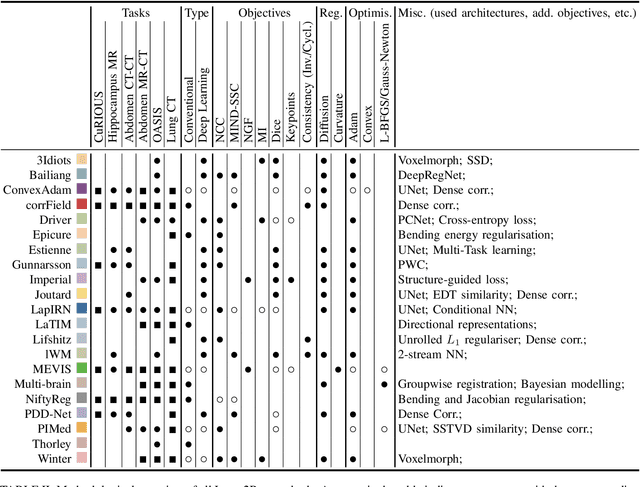
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
Deformable Image Registration with Deep Network Priors: a Study on Longitudinal PET Images
Nov 22, 2021



Abstract:Longitudinal image registration is challenging and has not yet benefited from major performance improvements thanks to deep-learning. Inspired by Deep Image Prior, this paper introduces a different use of deep architectures as regularizers to tackle the image registration question. We propose a subject-specific deformable registration method called MIRRBA, relying on a deep pyramidal architecture to be the prior parametric model constraining the deformation field. Diverging from the supervised learning paradigm, MIRRBA does not require a learning database, but only the pair of images to be registered to optimize the network's parameters and provide a deformation field. We demonstrate the regularizing power of deep architectures and present new elements to understand the role of the architecture in deep learning methods for registration. Hence, to study the impact of the network parameters, we ran our method with different architectural configurations on a private dataset of 110 metastatic breast cancer full-body PET images with manual segmentations of the brain, bladder and metastatic lesions. We compared it against conventional iterative registration approaches and supervised deep learning-based models. Global and local registration accuracies were evaluated using the detection rate and the Dice score respectively, while registration realism was evaluated using the Jacobian's determinant. Moreover, we computed the ability of the different methods to shrink vanishing lesions with the disappearing rate. MIRRBA significantly improves the organ and lesion Dice scores of supervised models. Regarding the disappearing rate, MIRRBA more than doubles the best performing conventional approach SyNCC score. Our work therefore proposes an alternative way to bridge the performance gap between conventional and deep learning-based methods and demonstrates the regularizing power of deep architectures.
Kidney tumor segmentation using an ensembling multi-stage deep learning approach. A contribution to the KiTS19 challenge
Sep 02, 2019



Abstract:Precise characterization of the kidney and kidney tumor characteristics is of outmost importance in the context of kidney cancer treatment, especially for nephron sparing surgery which requires a precise localization of the tissues to be removed. The need for accurate and automatic delineation tools is at the origin of the KiTS19 challenge. It aims at accelerating the research and development in this field to aid prognosis and treatment planning by providing a characterized dataset of 300 CT scans to be segmented. To address the challenge, we proposed an automatic, multi-stage, 2.5D deep learning-based segmentation approach based on Residual UNet framework. An ensembling operation is added at the end to combine prediction results from previous stages reducing the variance between single models. Our neural network segmentation algorithm reaches a mean Dice score of 0.96 and 0.74 for kidney and kidney tumors, respectively on 90 unseen test cases. The results obtained are promising and could be improved by incorporating prior knowledge about the benign cysts that regularly lower the tumor segmentation results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge