Albert C. S. Chung
More than Segmentation: Benchmarking SAM 3 for Segmentation, 3D Perception, and Reconstruction in Robotic Surgery
Dec 10, 2025Abstract:The recent SAM 3 and SAM 3D have introduced significant advancements over the predecessor, SAM 2, particularly with the integration of language-based segmentation and enhanced 3D perception capabilities. SAM 3 supports zero-shot segmentation across a wide range of prompts, including point, bounding box, and language-based prompts, allowing for more flexible and intuitive interactions with the model. In this empirical evaluation, we assess the performance of SAM 3 in robot-assisted surgery, benchmarking its zero-shot segmentation with point and bounding box prompts and exploring its effectiveness in dynamic video tracking, alongside its newly introduced language prompt segmentation. While language prompts show potential, their performance in the surgical domain is currently suboptimal, highlighting the need for further domain-specific training. Additionally, we investigate SAM 3D's depth reconstruction abilities, demonstrating its capacity to process surgical scene data and reconstruct 3D anatomical structures from 2D images. Through comprehensive testing on the MICCAI EndoVis 2017 and EndoVis 2018 benchmarks, SAM 3 shows clear improvements over SAM and SAM 2 in both image and video segmentation under spatial prompts, while the zero-shot evaluations of SAM 3D on SCARED, StereoMIS, and EndoNeRF indicate strong monocular depth estimation and realistic 3D instrument reconstruction, yet also reveal remaining limitations in complex, highly dynamic surgical scenes.
SETGen: Scalable and Efficient Template Generation Framework for Groupwise Medical Image Registration
Nov 10, 2022



Abstract:Template generation is a crucial step of groupwise image registration which deforms a group of subjects into a common space. Existing traditional and deep learning-based methods can generate high-quality template images. However, they suffer from substantial time costs or limited application scenarios like fixed group size. In this paper, we propose an efficient groupwise template generative framework based on variational autoencoder models utilizing the arithmetic property of latent representation of input images. We acquire the latent vectors of each input and use the average vector to construct the template through the decoder. Therefore, the method can be applied to groups of any scale. Secondly, we explore a siamese training scheme that feeds two images to the shared-weight twin networks and compares the distances between inputs and the generated template to prompt the template to be close to the implicit center. We conduct experiments on 3D brain MRI scans of groups of different sizes. Results show that our framework can achieve comparable and even better performance to baselines, with runtime decreased to seconds.
Unsupervised Deformable Image Registration with Absent Correspondences in Pre-operative and Post-Recurrence Brain Tumor MRI Scans
Jun 08, 2022



Abstract:Registration of pre-operative and post-recurrence brain images is often needed to evaluate the effectiveness of brain gliomas treatment. While recent deep learning-based deformable registration methods have achieved remarkable success with healthy brain images, most of them would be unable to accurately align images with pathologies due to the absent correspondences in the reference image. In this paper, we propose a deep learning-based deformable registration method that jointly estimates regions with absent correspondence and bidirectional deformation fields. A forward-backward consistency constraint is used to aid in the localization of the resection and recurrence region from voxels with absence correspondences in the two images. Results on 3D clinical data from the BraTS-Reg challenge demonstrate our method can improve image alignment compared to traditional and deep learning-based registration approaches with or without cost function masking strategy. The source code is available at https://github.com/cwmok/DIRAC.
Affine Medical Image Registration with Coarse-to-Fine Vision Transformer
Mar 30, 2022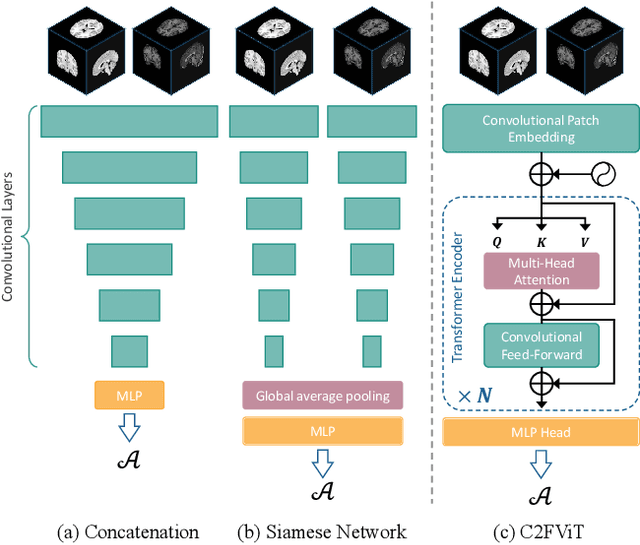
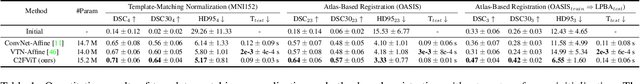
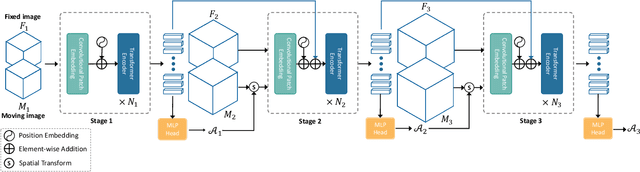
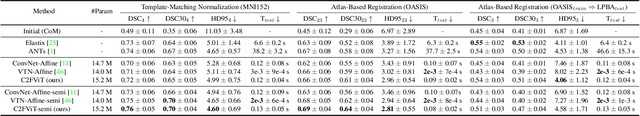
Abstract:Affine registration is indispensable in a comprehensive medical image registration pipeline. However, only a few studies focus on fast and robust affine registration algorithms. Most of these studies utilize convolutional neural networks (CNNs) to learn joint affine and non-parametric registration, while the standalone performance of the affine subnetwork is less explored. Moreover, existing CNN-based affine registration approaches focus either on the local misalignment or the global orientation and position of the input to predict the affine transformation matrix, which are sensitive to spatial initialization and exhibit limited generalizability apart from the training dataset. In this paper, we present a fast and robust learning-based algorithm, Coarse-to-Fine Vision Transformer (C2FViT), for 3D affine medical image registration. Our method naturally leverages the global connectivity and locality of the convolutional vision transformer and the multi-resolution strategy to learn the global affine registration. We evaluate our method on 3D brain atlas registration and template-matching normalization. Comprehensive results demonstrate that our method is superior to the existing CNNs-based affine registration methods in terms of registration accuracy, robustness and generalizability while preserving the runtime advantage of the learning-based methods. The source code is available at https://github.com/cwmok/C2FViT.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021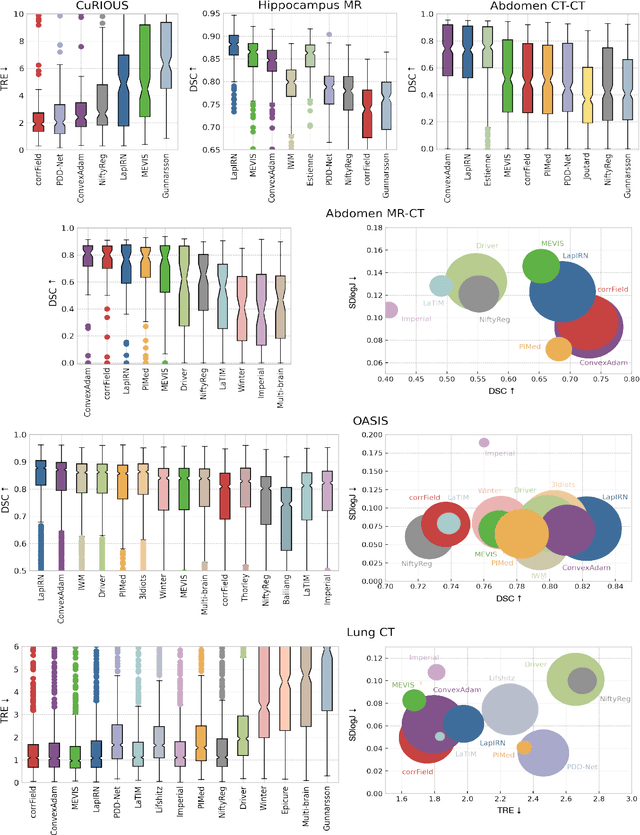
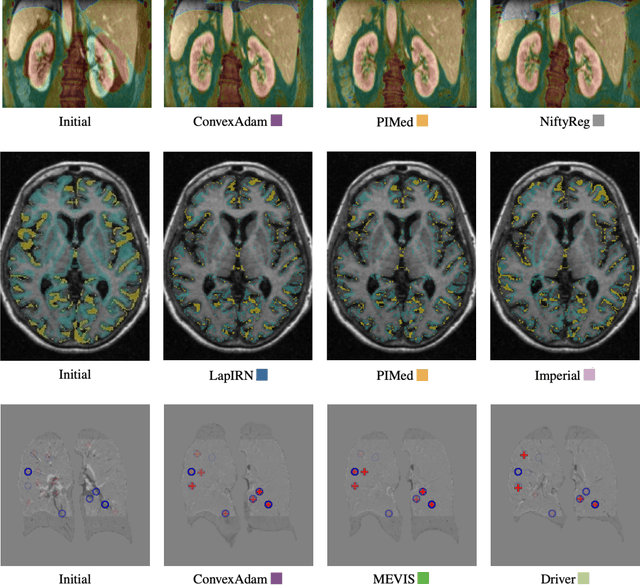
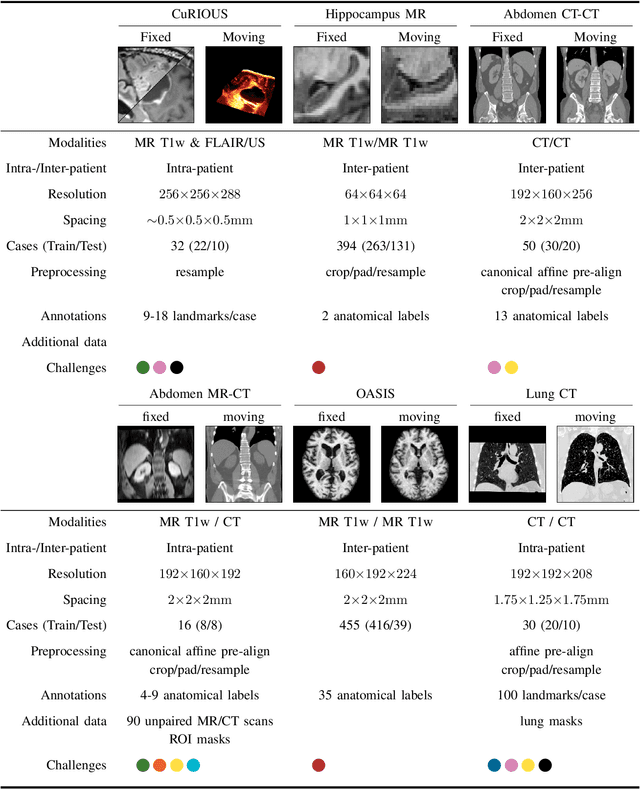
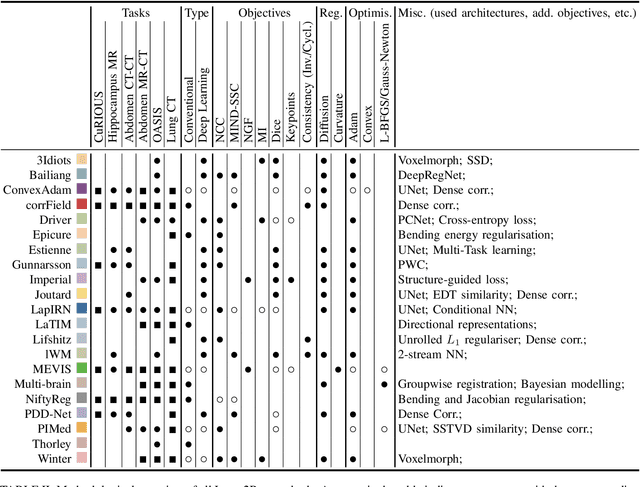
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
Conditional Deformable Image Registration with Convolutional Neural Network
Jul 05, 2021



Abstract:Recent deep learning-based methods have shown promising results and runtime advantages in deformable image registration. However, analyzing the effects of hyperparameters and searching for optimal regularization parameters prove to be too prohibitive in deep learning-based methods. This is because it involves training a substantial number of separate models with distinct hyperparameter values. In this paper, we propose a conditional image registration method and a new self-supervised learning paradigm for deep deformable image registration. By learning the conditional features that are correlated with the regularization hyperparameter, we demonstrate that optimal solutions with arbitrary hyperparameters can be captured by a single deep convolutional neural network. In addition, the smoothness of the resulting deformation field can be manipulated with arbitrary strength of smoothness regularization during inference. Extensive experiments on a large-scale brain MRI dataset show that our proposed method enables the precise control of the smoothness of the deformation field without sacrificing the runtime advantage or registration accuracy.
Edge-variational Graph Convolutional Networks for Uncertainty-aware Disease Prediction
Sep 06, 2020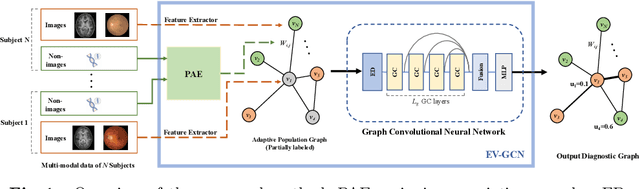
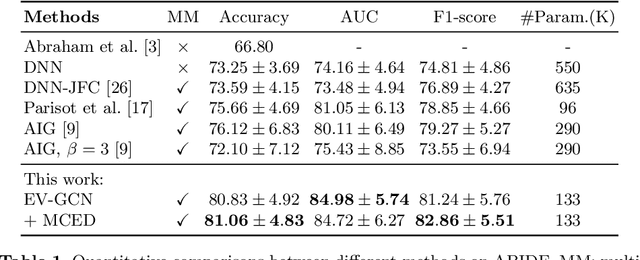
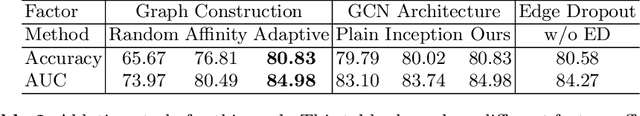
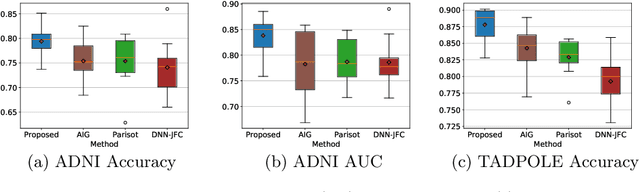
Abstract:There is a rising need for computational models that can complementarily leverage data of different modalities while investigating associations between subjects for population-based disease analysis. Despite the success of convolutional neural networks in representation learning for imaging data, it is still a very challenging task. In this paper, we propose a generalizable framework that can automatically integrate imaging data with non-imaging data in populations for uncertainty-aware disease prediction. At its core is a learnable adaptive population graph with variational edges, which we mathematically prove that it is optimizable in conjunction with graph convolutional neural networks. To estimate the predictive uncertainty related to the graph topology, we propose the novel concept of Monte-Carlo edge dropout. Experimental results on four databases show that our method can consistently and significantly improve the diagnostic accuracy for Autism spectrum disorder, Alzheimer's disease, and ocular diseases, indicating its generalizability in leveraging multimodal data for computer-aided diagnosis.
Large Deformation Diffeomorphic Image Registration with Laplacian Pyramid Networks
Jun 30, 2020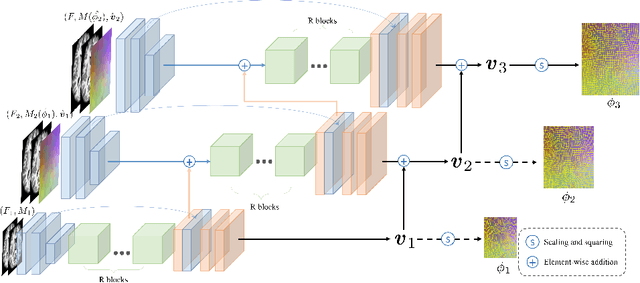
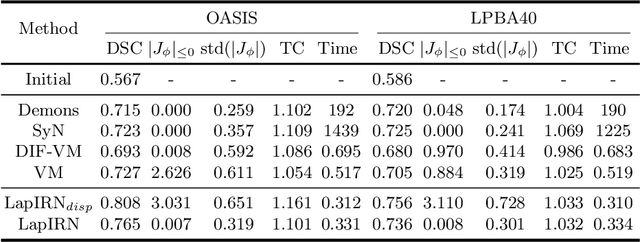
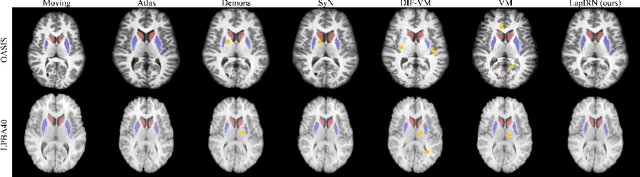
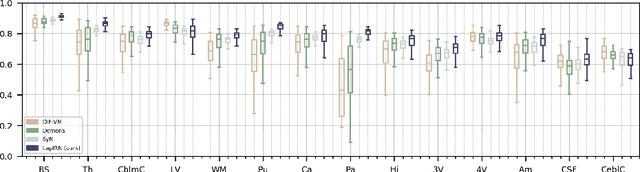
Abstract:Deep learning-based methods have recently demonstrated promising results in deformable image registration for a wide range of medical image analysis tasks. However, existing deep learning-based methods are usually limited to small deformation settings, and desirable properties of the transformation including bijective mapping and topology preservation are often being ignored by these approaches. In this paper, we propose a deep Laplacian Pyramid Image Registration Network, which can solve the image registration optimization problem in a coarse-to-fine fashion within the space of diffeomorphic maps. Extensive quantitative and qualitative evaluations on two MR brain scan datasets show that our method outperforms the existing methods by a significant margin while maintaining desirable diffeomorphic properties and promising registration speed.
Fast Symmetric Diffeomorphic Image Registration with Convolutional Neural Networks
Mar 20, 2020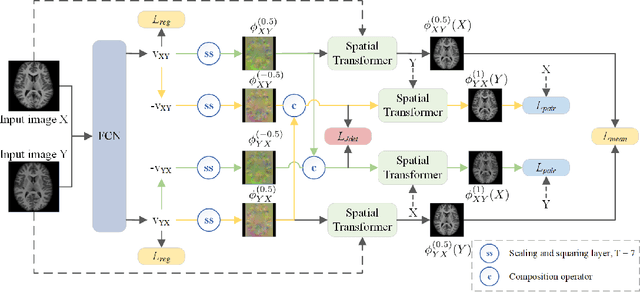
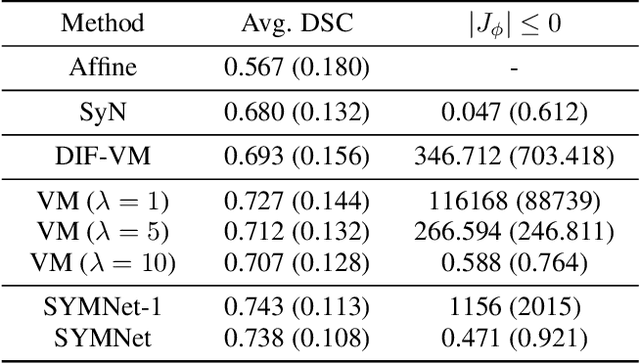
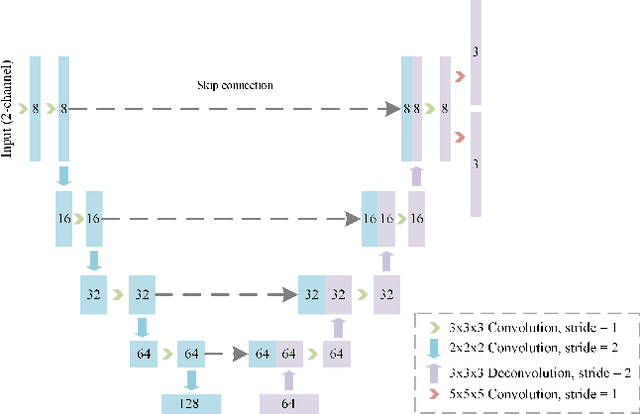
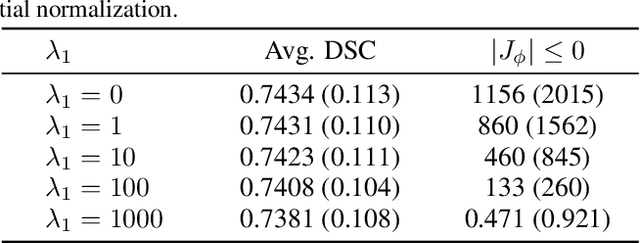
Abstract:Diffeomorphic deformable image registration is crucial in many medical image studies, as it offers unique, special properties including topology preservation and invertibility of the transformation. Recent deep learning-based deformable image registration methods achieve fast image registration by leveraging a convolutional neural network (CNN) to learn the spatial transformation from the synthetic ground truth or the similarity metric. However, these approaches often ignore the topology preservation of the transformation and the smoothness of the transformation which is enforced by a global smoothing energy function alone. Moreover, deep learning-based approaches often estimate the displacement field directly, which cannot guarantee the existence of the inverse transformation. In this paper, we present a novel, efficient unsupervised symmetric image registration method which maximizes the similarity between images within the space of diffeomorphic maps and estimates both forward and inverse transformations simultaneously. We evaluate our method on 3D image registration with a large scale brain image dataset. Our method achieves state-of-the-art registration accuracy and running time while maintaining desirable diffeomorphic properties.
CELNet: Evidence Localization for Pathology Images using Weakly Supervised Learning
Sep 16, 2019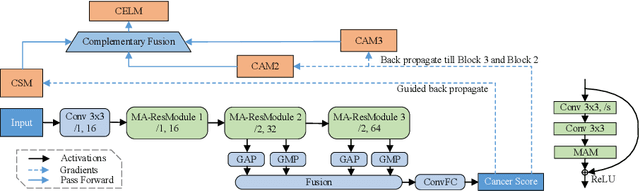
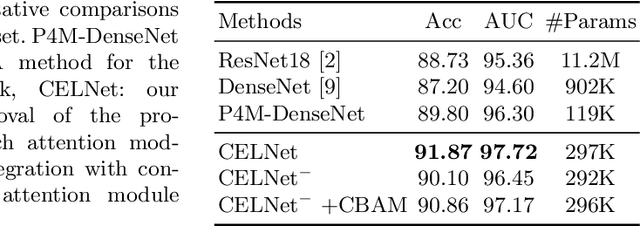
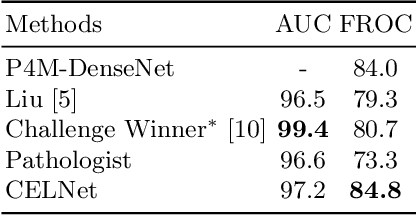
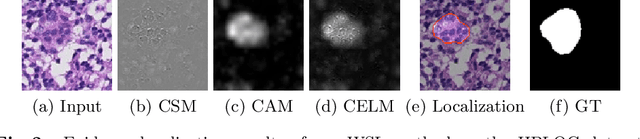
Abstract:Despite deep convolutional neural networks boost the performance of image classification and segmentation in digital pathology analysis, they are usually weak in interpretability for clinical applications or require heavy annotations to achieve object localization. To overcome this problem, we propose a weakly supervised learning-based approach that can effectively learn to localize the discriminative evidence for a diagnostic label from weakly labeled training data. Experimental results show that our proposed method can reliably pinpoint the location of cancerous evidence supporting the decision of interest, while still achieving a competitive performance on glimpse-level and slide-level histopathologic cancer detection tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge