Yongxiang Huang
CannyEdit: Selective Canny Control and Dual-Prompt Guidance for Training-Free Image Editing
Aug 09, 2025Abstract:Recent advances in text-to-image (T2I) models have enabled training-free regional image editing by leveraging the generative priors of foundation models. However, existing methods struggle to balance text adherence in edited regions, context fidelity in unedited areas, and seamless integration of edits. We introduce CannyEdit, a novel training-free framework that addresses these challenges through two key innovations: (1) Selective Canny Control, which masks the structural guidance of Canny ControlNet in user-specified editable regions while strictly preserving details of the source images in unedited areas via inversion-phase ControlNet information retention. This enables precise, text-driven edits without compromising contextual integrity. (2) Dual-Prompt Guidance, which combines local prompts for object-specific edits with a global target prompt to maintain coherent scene interactions. On real-world image editing tasks (addition, replacement, removal), CannyEdit outperforms prior methods like KV-Edit, achieving a 2.93 to 10.49 percent improvement in the balance of text adherence and context fidelity. In terms of editing seamlessness, user studies reveal only 49.2 percent of general users and 42.0 percent of AIGC experts identified CannyEdit's results as AI-edited when paired with real images without edits, versus 76.08 to 89.09 percent for competitor methods.
Dual Risk Minimization: Towards Next-Level Robustness in Fine-tuning Zero-Shot Models
Nov 29, 2024



Abstract:Fine-tuning foundation models often compromises their robustness to distribution shifts. To remedy this, most robust fine-tuning methods aim to preserve the pre-trained features. However, not all pre-trained features are robust and those methods are largely indifferent to which ones to preserve. We propose dual risk minimization (DRM), which combines empirical risk minimization with worst-case risk minimization, to better preserve the core features of downstream tasks. In particular, we utilize core-feature descriptions generated by LLMs to induce core-based zero-shot predictions which then serve as proxies to estimate the worst-case risk. DRM balances two crucial aspects of model robustness: expected performance and worst-case performance, establishing a new state of the art on various real-world benchmarks. DRM significantly improves the out-of-distribution performance of CLIP ViT-L/14@336 on ImageNet (75.9 to 77.1), WILDS-iWildCam (47.1 to 51.8), and WILDS-FMoW (50.7 to 53.1); opening up new avenues for robust fine-tuning. Our code is available at https://github.com/vaynexie/DRM .
Towards Fine-Grained Explainability for Heterogeneous Graph Neural Network
Dec 23, 2023Abstract:Heterogeneous graph neural networks (HGNs) are prominent approaches to node classification tasks on heterogeneous graphs. Despite the superior performance, insights about the predictions made from HGNs are obscure to humans. Existing explainability techniques are mainly proposed for GNNs on homogeneous graphs. They focus on highlighting salient graph objects to the predictions whereas the problem of how these objects affect the predictions remains unsolved. Given heterogeneous graphs with complex structures and rich semantics, it is imperative that salient objects can be accompanied with their influence paths to the predictions, unveiling the reasoning process of HGNs. In this paper, we develop xPath, a new framework that provides fine-grained explanations for black-box HGNs specifying a cause node with its influence path to the target node. In xPath, we differentiate the influence of a node on the prediction w.r.t. every individual influence path, and measure the influence by perturbing graph structure via a novel graph rewiring algorithm. Furthermore, we introduce a greedy search algorithm to find the most influential fine-grained explanations efficiently. Empirical results on various HGNs and heterogeneous graphs show that xPath yields faithful explanations efficiently, outperforming the adaptations of advanced GNN explanation approaches.
A Causal Framework to Unify Common Domain Generalization Approaches
Jul 13, 2023



Abstract:Domain generalization (DG) is about learning models that generalize well to new domains that are related to, but different from, the training domain(s). It is a fundamental problem in machine learning and has attracted much attention in recent years. A large number of approaches have been proposed. Different approaches are motivated from different perspectives, making it difficult to gain an overall understanding of the area. In this paper, we propose a causal framework for domain generalization and present an understanding of common DG approaches in the framework. Our work sheds new lights on the following questions: (1) What are the key ideas behind each DG method? (2) Why is it expected to improve generalization to new domains theoretically? (3) How are different DG methods related to each other and what are relative advantages and limitations? By providing a unified perspective on DG, we hope to help researchers better understand the underlying principles and develop more effective approaches for this critical problem in machine learning.
Model Debiasing via Gradient-based Explanation on Representation
May 20, 2023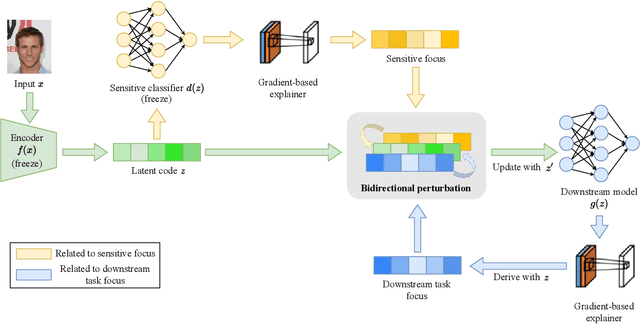

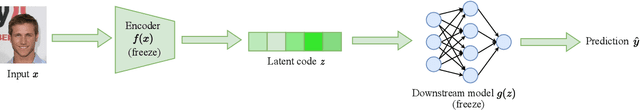

Abstract:Machine learning systems produce biased results towards certain demographic groups, known as the fairness problem. Recent approaches to tackle this problem learn a latent code (i.e., representation) through disentangled representation learning and then discard the latent code dimensions correlated with sensitive attributes (e.g., gender). Nevertheless, these approaches may suffer from incomplete disentanglement and overlook proxy attributes (proxies for sensitive attributes) when processing real-world data, especially for unstructured data, causing performance degradation in fairness and loss of useful information for downstream tasks. In this paper, we propose a novel fairness framework that performs debiasing with regard to both sensitive attributes and proxy attributes, which boosts the prediction performance of downstream task models without complete disentanglement. The main idea is to, first, leverage gradient-based explanation to find two model focuses, 1) one focus for predicting sensitive attributes and 2) the other focus for predicting downstream task labels, and second, use them to perturb the latent code that guides the training of downstream task models towards fairness and utility goals. We show empirically that our framework works with both disentangled and non-disentangled representation learning methods and achieves better fairness-accuracy trade-off on unstructured and structured datasets than previous state-of-the-art approaches.
Contrastive Domain Generalization via Logit Attribution Matching
May 13, 2023Abstract:Domain Generalization (DG) is an important open problem in machine learning. Deep models are susceptible to domain shifts of even minute degrees, which severely compromises their reliability in real applications. To alleviate the issue, most existing methods enforce various invariant constraints across multiple training domains. However,such an approach provides little performance guarantee for novel test domains in general. In this paper, we investigate a different approach named Contrastive Domain Generalization (CDG), which exploits semantic invariance exhibited by strongly contrastive data pairs in lieu of multiple domains. We present a causal DG theory that shows the potential capability of CDG; together with a regularization technique, Logit Attribution Matching (LAM), for realizing CDG. We empirically show that LAM outperforms state-of-the-art DG methods with only a small portion of paired data and that LAM helps models better focus on semantic features which are crucial to DG.
Edge-variational Graph Convolutional Networks for Uncertainty-aware Disease Prediction
Sep 06, 2020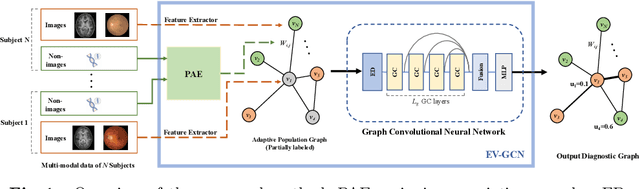
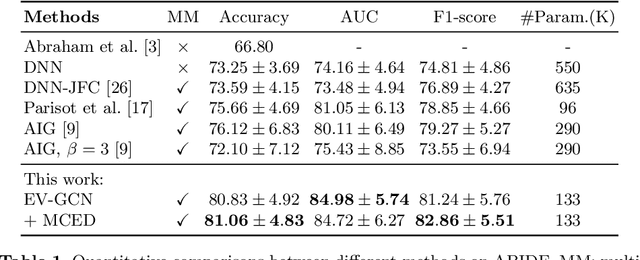
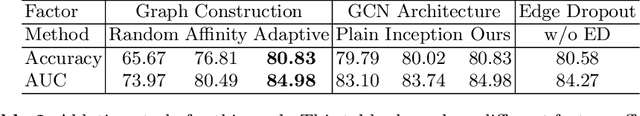
Abstract:There is a rising need for computational models that can complementarily leverage data of different modalities while investigating associations between subjects for population-based disease analysis. Despite the success of convolutional neural networks in representation learning for imaging data, it is still a very challenging task. In this paper, we propose a generalizable framework that can automatically integrate imaging data with non-imaging data in populations for uncertainty-aware disease prediction. At its core is a learnable adaptive population graph with variational edges, which we mathematically prove that it is optimizable in conjunction with graph convolutional neural networks. To estimate the predictive uncertainty related to the graph topology, we propose the novel concept of Monte-Carlo edge dropout. Experimental results on four databases show that our method can consistently and significantly improve the diagnostic accuracy for Autism spectrum disorder, Alzheimer's disease, and ocular diseases, indicating its generalizability in leveraging multimodal data for computer-aided diagnosis.
CELNet: Evidence Localization for Pathology Images using Weakly Supervised Learning
Sep 16, 2019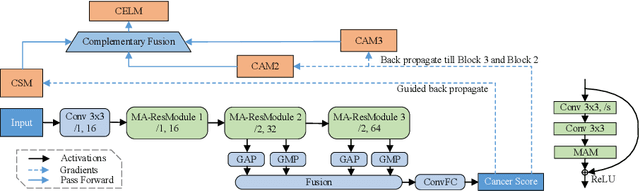
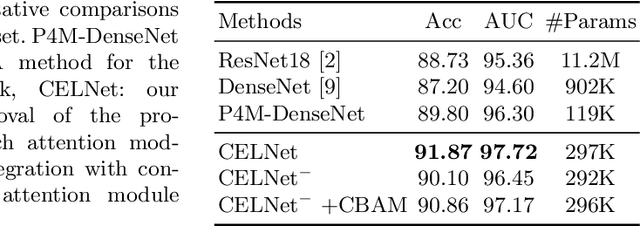
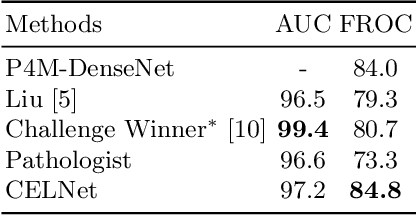
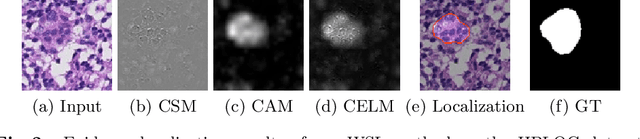
Abstract:Despite deep convolutional neural networks boost the performance of image classification and segmentation in digital pathology analysis, they are usually weak in interpretability for clinical applications or require heavy annotations to achieve object localization. To overcome this problem, we propose a weakly supervised learning-based approach that can effectively learn to localize the discriminative evidence for a diagnostic label from weakly labeled training data. Experimental results show that our proposed method can reliably pinpoint the location of cancerous evidence supporting the decision of interest, while still achieving a competitive performance on glimpse-level and slide-level histopathologic cancer detection tasks.
Improving High Resolution Histology Image Classification with Deep Spatial Fusion Network
Jul 27, 2018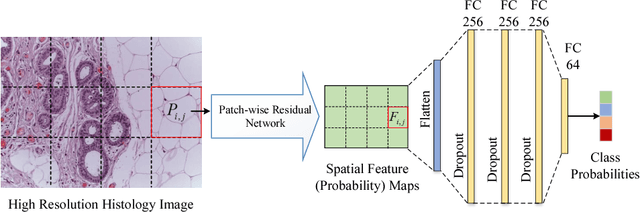
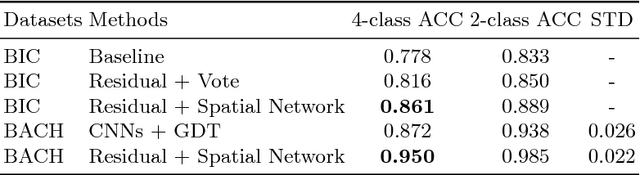
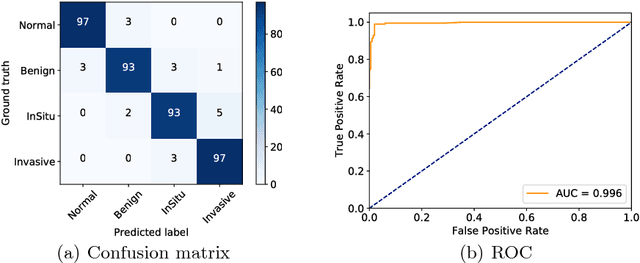
Abstract:Histology imaging is an essential diagnosis method to finalize the grade and stage of cancer of different tissues, especially for breast cancer diagnosis. Specialists often disagree on the final diagnosis on biopsy tissue due to the complex morphological variety. Although convolutional neural networks (CNN) have advantages in extracting discriminative features in image classification, directly training a CNN on high resolution histology images is computationally infeasible currently. Besides, inconsistent discriminative features often distribute over the whole histology image, which incurs challenges in patch-based CNN classification method. In this paper, we propose a novel architecture for automatic classification of high resolution histology images. First, an adapted residual network is employed to explore hierarchical features without attenuation. Second, we develop a robust deep fusion network to utilize the spatial relationship between patches and learn to correct the prediction bias generated from inconsistent discriminative feature distribution. The proposed method is evaluated using 10-fold cross-validation on 400 high resolution breast histology images with balanced labels and reports 95% accuracy on 4-class classification and 98.5% accuracy, 99.6% AUC on 2-class classification (carcinoma and non-carcinoma), which substantially outperforms previous methods and close to pathologist performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge