Rongzhao Zhang
DynaWeb: Model-Based Reinforcement Learning of Web Agents
Jan 29, 2026Abstract:The development of autonomous web agents, powered by Large Language Models (LLMs) and reinforcement learning (RL), represents a significant step towards general-purpose AI assistants. However, training these agents is severely hampered by the challenges of interacting with the live internet, which is inefficient, costly, and fraught with risks. Model-based reinforcement learning (MBRL) offers a promising solution by learning a world model of the environment to enable simulated interaction. This paper introduces DynaWeb, a novel MBRL framework that trains web agents through interacting with a web world model trained to predict naturalistic web page representations given agent actions. This model serves as a synthetic web environment where an agent policy can dream by generating vast quantities of rollout action trajectories for efficient online reinforcement learning. Beyond free policy rollouts, DynaWeb incorporates real expert trajectories from training data, which are randomly interleaved with on-policy rollouts during training to improve stability and sample efficiency. Experiments conducted on the challenging WebArena and WebVoyager benchmarks demonstrate that DynaWeb consistently and significantly improves the performance of state-of-the-art open-source web agent models. Our findings establish the viability of training web agents through imagination, offering a scalable and efficient way to scale up online agentic RL.
Multi-Agent Intelligence for Multidisciplinary Decision-Making in Gastrointestinal Oncology
Dec 23, 2025Abstract:Multimodal clinical reasoning in the field of gastrointestinal (GI) oncology necessitates the integrated interpretation of endoscopic imagery, radiological data, and biochemical markers. Despite the evident potential exhibited by Multimodal Large Language Models (MLLMs), they frequently encounter challenges such as context dilution and hallucination when confronted with intricate, heterogeneous medical histories. In order to address these limitations, a hierarchical Multi-Agent Framework is proposed, which emulates the collaborative workflow of a human Multidisciplinary Team (MDT). The system attained a composite expert evaluation score of 4.60/5.00, thereby demonstrating a substantial improvement over the monolithic baseline. It is noteworthy that the agent-based architecture yielded the most substantial enhancements in reasoning logic and medical accuracy. The findings indicate that mimetic, agent-based collaboration provides a scalable, interpretable, and clinically robust paradigm for automated decision support in oncology.
MedBench v4: A Robust and Scalable Benchmark for Evaluating Chinese Medical Language Models, Multimodal Models, and Intelligent Agents
Nov 19, 2025Abstract:Recent advances in medical large language models (LLMs), multimodal models, and agents demand evaluation frameworks that reflect real clinical workflows and safety constraints. We present MedBench v4, a nationwide, cloud-based benchmarking infrastructure comprising over 700,000 expert-curated tasks spanning 24 primary and 91 secondary specialties, with dedicated tracks for LLMs, multimodal models, and agents. Items undergo multi-stage refinement and multi-round review by clinicians from more than 500 institutions, and open-ended responses are scored by an LLM-as-a-judge calibrated to human ratings. We evaluate 15 frontier models. Base LLMs reach a mean overall score of 54.1/100 (best: Claude Sonnet 4.5, 62.5/100), but safety and ethics remain low (18.4/100). Multimodal models perform worse overall (mean 47.5/100; best: GPT-5, 54.9/100), with solid perception yet weaker cross-modal reasoning. Agents built on the same backbones substantially improve end-to-end performance (mean 79.8/100), with Claude Sonnet 4.5-based agents achieving up to 85.3/100 overall and 88.9/100 on safety tasks. MedBench v4 thus reveals persisting gaps in multimodal reasoning and safety for base models, while showing that governance-aware agentic orchestration can markedly enhance benchmarked clinical readiness without sacrificing capability. By aligning tasks with Chinese clinical guidelines and regulatory priorities, the platform offers a practical reference for hospitals, developers, and policymakers auditing medical AI.
Benchmarking Ethical and Safety Risks of Healthcare LLMs in China-Toward Systemic Governance under Healthy China 2030
May 12, 2025
Abstract:Large Language Models (LLMs) are poised to transform healthcare under China's Healthy China 2030 initiative, yet they introduce new ethical and patient-safety challenges. We present a novel 12,000-item Q&A benchmark covering 11 ethics and 9 safety dimensions in medical contexts, to quantitatively evaluate these risks. Using this dataset, we assess state-of-the-art Chinese medical LLMs (e.g., Qwen 2.5-32B, DeepSeek), revealing moderate baseline performance (accuracy 42.7% for Qwen 2.5-32B) and significant improvements after fine-tuning on our data (up to 50.8% accuracy). Results show notable gaps in LLM decision-making on ethics and safety scenarios, reflecting insufficient institutional oversight. We then identify systemic governance shortfalls-including the lack of fine-grained ethical audit protocols, slow adaptation by hospital IRBs, and insufficient evaluation tools-that currently hinder safe LLM deployment. Finally, we propose a practical governance framework for healthcare institutions (embedding LLM auditing teams, enacting data ethics guidelines, and implementing safety simulation pipelines) to proactively manage LLM risks. Our study highlights the urgent need for robust LLM governance in Chinese healthcare, aligning AI innovation with patient safety and ethical standards.
Towards All-in-One Medical Image Re-Identification
Mar 11, 2025Abstract:Medical image re-identification (MedReID) is under-explored so far, despite its critical applications in personalized healthcare and privacy protection. In this paper, we introduce a thorough benchmark and a unified model for this problem. First, to handle various medical modalities, we propose a novel Continuous Modality-based Parameter Adapter (ComPA). ComPA condenses medical content into a continuous modality representation and dynamically adjusts the modality-agnostic model with modality-specific parameters at runtime. This allows a single model to adaptively learn and process diverse modality data. Furthermore, we integrate medical priors into our model by aligning it with a bag of pre-trained medical foundation models, in terms of the differential features. Compared to single-image feature, modeling the inter-image difference better fits the re-identification problem, which involves discriminating multiple images. We evaluate the proposed model against 25 foundation models and 8 large multi-modal language models across 11 image datasets, demonstrating consistently superior performance. Additionally, we deploy the proposed MedReID technique to two real-world applications, i.e., history-augmented personalized diagnosis and medical privacy protection. Codes and model is available at \href{https://github.com/tianyuan168326/All-in-One-MedReID-Pytorch}{https://github.com/tianyuan168326/All-in-One-MedReID-Pytorch}.
CAT: Coordinating Anatomical-Textual Prompts for Multi-Organ and Tumor Segmentation
Jun 11, 2024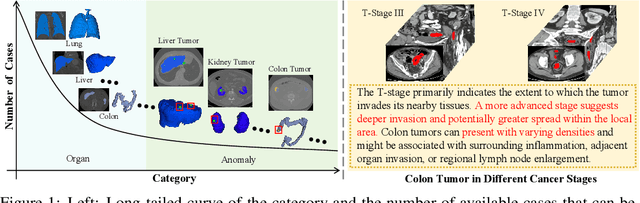
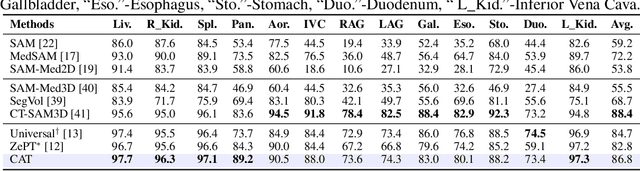
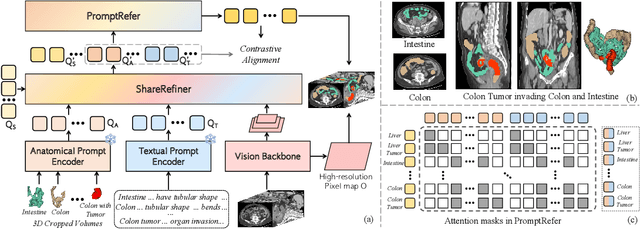
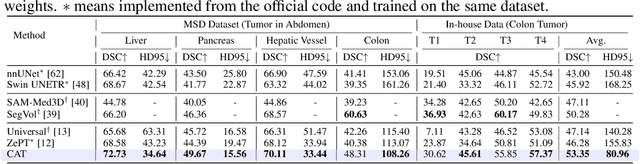
Abstract:Existing promptable segmentation methods in the medical imaging field primarily consider either textual or visual prompts to segment relevant objects, yet they often fall short when addressing anomalies in medical images, like tumors, which may vary greatly in shape, size, and appearance. Recognizing the complexity of medical scenarios and the limitations of textual or visual prompts, we propose a novel dual-prompt schema that leverages the complementary strengths of visual and textual prompts for segmenting various organs and tumors. Specifically, we introduce CAT, an innovative model that Coordinates Anatomical prompts derived from 3D cropped images with Textual prompts enriched by medical domain knowledge. The model architecture adopts a general query-based design, where prompt queries facilitate segmentation queries for mask prediction. To synergize two types of prompts within a unified framework, we implement a ShareRefiner, which refines both segmentation and prompt queries while disentangling the two types of prompts. Trained on a consortium of 10 public CT datasets, CAT demonstrates superior performance in multiple segmentation tasks. Further validation on a specialized in-house dataset reveals the remarkable capacity of segmenting tumors across multiple cancer stages. This approach confirms that coordinating multimodal prompts is a promising avenue for addressing complex scenarios in the medical domain.
GuideGen: A Text-guided Framework for Joint CT Volume and Anatomical structure Generation
Mar 12, 2024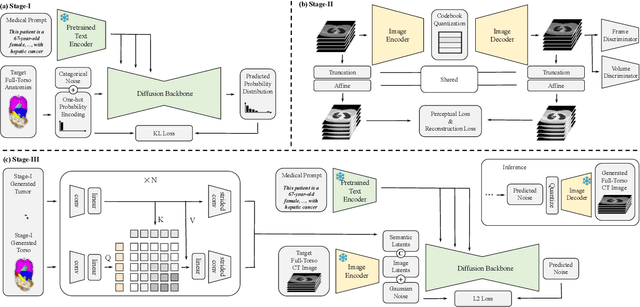
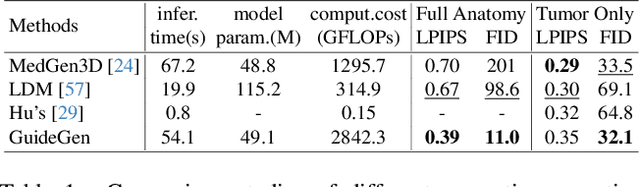
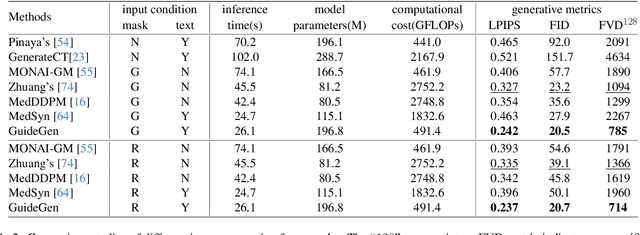
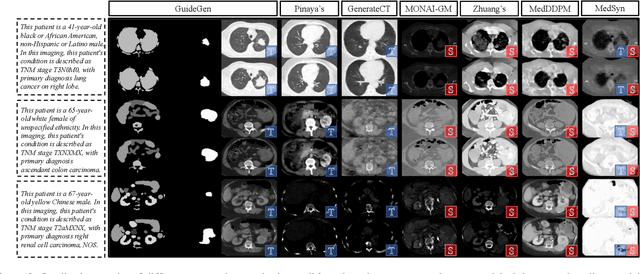
Abstract:The annotation burden and extensive labor for gathering a large medical dataset with images and corresponding labels are rarely cost-effective and highly intimidating. This results in a lack of abundant training data that undermines downstream tasks and partially contributes to the challenge image analysis faces in the medical field. As a workaround, given the recent success of generative neural models, it is now possible to synthesize image datasets at a high fidelity guided by external constraints. This paper explores this possibility and presents \textbf{GuideGen}: a pipeline that jointly generates CT images and tissue masks for abdominal organs and colorectal cancer conditioned on a text prompt. Firstly, we introduce Volumetric Mask Sampler to fit the discrete distribution of mask labels and generate low-resolution 3D tissue masks. Secondly, our Conditional Image Generator autoregressively generates CT slices conditioned on a corresponding mask slice to incorporate both style information and anatomical guidance. This pipeline guarantees high fidelity and variability as well as exact alignment between generated CT volumes and tissue masks. Both qualitative and quantitative experiments on 3D abdominal CTs demonstrate a high performance of our proposed pipeline, thereby proving our method can serve as a dataset generator and provide potential benefits to downstream tasks. It is hoped that our work will offer a promising solution on the multimodality generation of CT and its anatomical mask. Our source code is publicly available at https://github.com/OvO1111/JointImageGeneration.
ZePT: Zero-Shot Pan-Tumor Segmentation via Query-Disentangling and Self-Prompting
Dec 07, 2023



Abstract:The long-tailed distribution problem in medical image analysis reflects a high prevalence of common conditions and a low prevalence of rare ones, which poses a significant challenge in developing a unified model capable of identifying rare or novel tumor categories not encountered during training. In this paper, we propose a new zero-shot pan-tumor segmentation framework (ZePT) based on query-disentangling and self-prompting to segment unseen tumor categories beyond the training set. ZePT disentangles the object queries into two subsets and trains them in two stages. Initially, it learns a set of fundamental queries for organ segmentation through an object-aware feature grouping strategy, which gathers organ-level visual features. Subsequently, it refines the other set of advanced queries that focus on the auto-generated visual prompts for unseen tumor segmentation. Moreover, we introduce query-knowledge alignment at the feature level to enhance each query's discriminative representation and generalizability. Extensive experiments on various tumor segmentation tasks demonstrate the performance superiority of ZePT, which surpasses the previous counterparts and evidence the promising ability for zero-shot tumor segmentation in real-world settings. Codes will be made publicly available.
AG-CRC: Anatomy-Guided Colorectal Cancer Segmentation in CT with Imperfect Anatomical Knowledge
Oct 07, 2023



Abstract:When delineating lesions from medical images, a human expert can always keep in mind the anatomical structure behind the voxels. However, although high-quality (though not perfect) anatomical information can be retrieved from computed tomography (CT) scans with modern deep learning algorithms, it is still an open problem how these automatically generated organ masks can assist in addressing challenging lesion segmentation tasks, such as the segmentation of colorectal cancer (CRC). In this paper, we develop a novel Anatomy-Guided segmentation framework to exploit the auto-generated organ masks to aid CRC segmentation from CT, namely AG-CRC. First, we obtain multi-organ segmentation (MOS) masks with existing MOS models (e.g., TotalSegmentor) and further derive a more robust organ of interest (OOI) mask that may cover most of the colon-rectum and CRC voxels. Then, we propose an anatomy-guided training patch sampling strategy by optimizing a heuristic gain function that considers both the proximity of important regions (e.g., the tumor or organs of interest) and sample diversity. Third, we design a novel self-supervised learning scheme inspired by the topology of tubular organs like the colon to boost the model performance further. Finally, we employ a masked loss scheme to guide the model to focus solely on the essential learning region. We extensively evaluate the proposed method on two CRC segmentation datasets, where substantial performance improvement (5% to 9% in Dice) is achieved over current state-of-the-art medical image segmentation models, and the ablation studies further evidence the efficacy of every proposed component.
A Fine-Grain Error Map Prediction and Segmentation Quality Assessment Framework for Whole-Heart Segmentation
Jul 29, 2019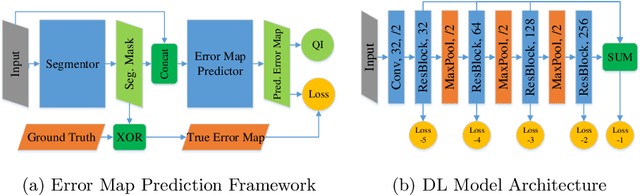
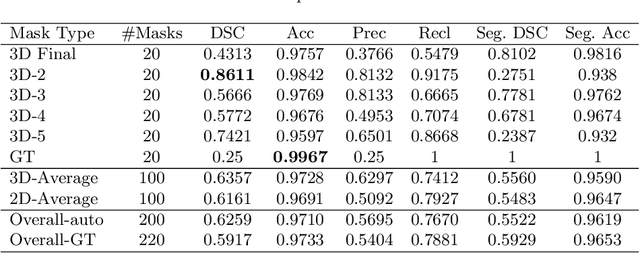
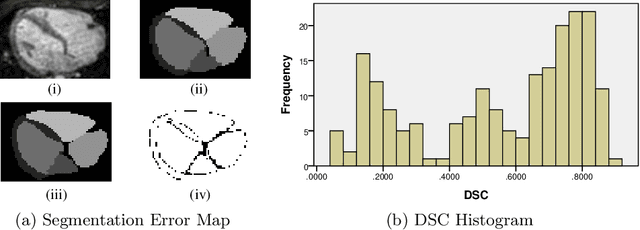
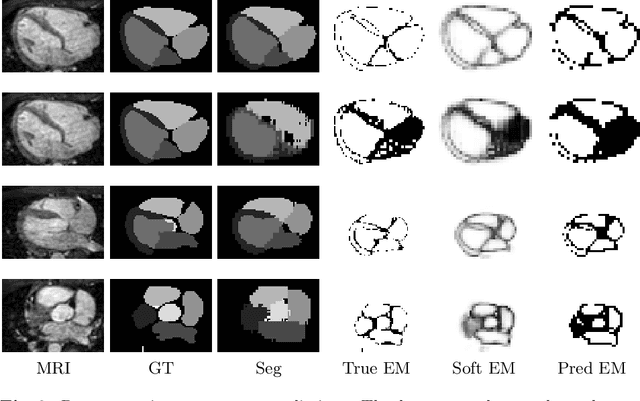
Abstract:When introducing advanced image computing algorithms, e.g., whole-heart segmentation, into clinical practice, a common suspicion is how reliable the automatically computed results are. In fact, it is important to find out the failure cases and identify the misclassified pixels so that they can be excluded or corrected for the subsequent analysis or diagnosis. However, it is not a trivial problem to predict the errors in a segmentation mask when ground truth (usually annotated by experts) is absent. In this work, we attempt to address the pixel-wise error map prediction problem and the per-case mask quality assessment problem using a unified deep learning (DL) framework. Specifically, we first formalize an error map prediction problem, then we convert it to a segmentation problem and build a DL network to tackle it. We also derive a quality indicator (QI) from a predicted error map to measure the overall quality of a segmentation mask. To evaluate the proposed framework, we perform extensive experiments on a public whole-heart segmentation dataset, i.e., MICCAI 2017 MMWHS. By 5-fold cross validation, we obtain an overall Dice score of 0.626 for the error map prediction task, and observe a high Pearson correlation coefficient (PCC) of 0.972 between QI and the actual segmentation accuracy (Acc), as well as a low mean absolute error (MAE) of 0.0048 between them, which evidences the efficacy of our method in both error map prediction and quality assessment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge