Lifeng Zhu
MRUCT: Mixed Reality Assistance for Acupuncture Guided by Ultrasonic Computed Tomography
Feb 12, 2025Abstract:Chinese acupuncture practitioners primarily depend on muscle memory and tactile feedback to insert needles and accurately target acupuncture points, as the current workflow lacks imaging modalities and visual aids. Consequently, new practitioners often learn through trial and error, requiring years of experience to become proficient and earn the trust of patients. Medical students face similar challenges in mastering this skill. To address these challenges, we developed an innovative system, MRUCT, that integrates ultrasonic computed tomography (UCT) with mixed reality (MR) technology to visualize acupuncture points in real-time. This system offers offline image registration and real-time guidance during needle insertion, enabling them to accurately position needles based on anatomical structures such as bones, muscles, and auto-generated reference points, with the potential for clinical implementation. In this paper, we outline the non-rigid registration methods used to reconstruct anatomical structures from UCT data, as well as the key design considerations of the MR system. We evaluated two different 3D user interface (3DUI) designs and compared the performance of our system to traditional workflows for both new practitioners and medical students. The results highlight the potential of MR to enhance therapeutic medical practices and demonstrate the effectiveness of the system we developed.
Facial Surgery Preview Based on the Orthognathic Treatment Prediction
Dec 15, 2024



Abstract:Orthognathic surgery consultation is essential to help patients understand the changes to their facial appearance after surgery. However, current visualization methods are often inefficient and inaccurate due to limited pre- and post-treatment data and the complexity of the treatment. To overcome these challenges, this study aims to develop a fully automated pipeline that generates accurate and efficient 3D previews of postsurgical facial appearances for patients with orthognathic treatment without requiring additional medical images. The study introduces novel aesthetic losses, such as mouth-convexity and asymmetry losses, to improve the accuracy of facial surgery prediction. Additionally, it proposes a specialized parametric model for 3D reconstruction of the patient, medical-related losses to guide latent code prediction network optimization, and a data augmentation scheme to address insufficient data. The study additionally employs FLAME, a parametric model, to enhance the quality of facial appearance previews by extracting facial latent codes and establishing dense correspondences between pre- and post-surgery geometries. Quantitative comparisons showed the algorithm's effectiveness, and qualitative results highlighted accurate facial contour and detail predictions. A user study confirmed that doctors and the public could not distinguish between machine learning predictions and actual postoperative results. This study aims to offer a practical, effective solution for orthognathic surgery consultations, benefiting doctors and patients.
DipMe: Haptic Recognition of Granular Media for Tangible Interactive Applications
Nov 13, 2024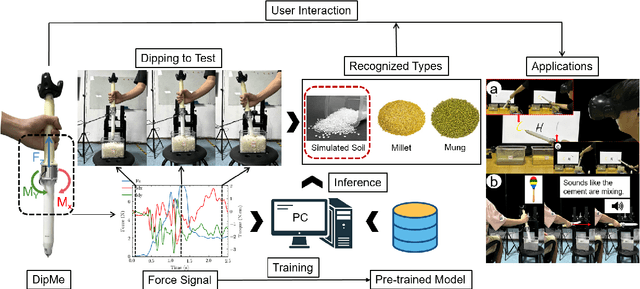

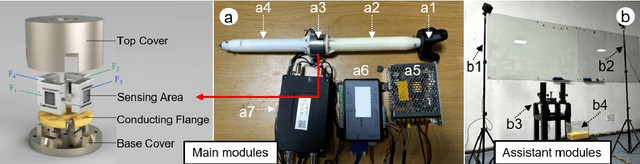
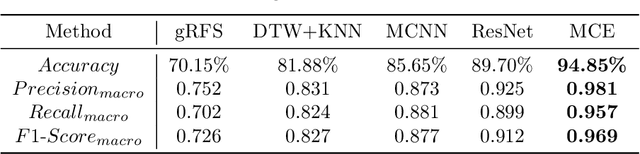
Abstract:While tangible user interface has shown its power in naturally interacting with rigid or soft objects, users cannot conveniently use different types of granular materials as the interaction media. We introduce DipMe as a smart device to recognize the types of granular media in real time, which can be used to connect the granular materials in the physical world with various virtual content. Other than vision-based solutions, we propose a dip operation of our device and exploit the haptic signals to recognize different types of granular materials. With modern machine learning tools, we find the haptic signals from different granular media are distinguishable by DipMe. With the online granular object recognition, we build several tangible interactive applications, demonstrating the effects of DipMe in perceiving granular materials and its potential in developing a tangible user interface with granular objects as the new media.
MSDiagnosis: An EMR-based Dataset for Clinical Multi-Step Diagnosis
Aug 19, 2024



Abstract:Clinical diagnosis is critical in medical practice, typically requiring a continuous and evolving process that includes primary diagnosis, differential diagnosis, and final diagnosis. However, most existing clinical diagnostic tasks are single-step processes, which does not align with the complex multi-step diagnostic procedures found in real-world clinical settings. In this paper, we propose a multi-step diagnostic task and annotate a clinical diagnostic dataset (MSDiagnosis). This dataset includes primary diagnosis, differential diagnosis, and final diagnosis questions. Additionally, we propose a novel and effective framework. This framework combines forward inference, backward inference, reflection, and refinement, enabling the LLM to self-evaluate and adjust its diagnostic results. To assess the effectiveness of our proposed method, we design and conduct extensive experiments. The experimental results demonstrate the effectiveness of the proposed method. We also provide a comprehensive experimental analysis and suggest future research directions for this task.
Cost-effective Instruction Learning for Pathology Vision and Language Analysis
Jul 25, 2024
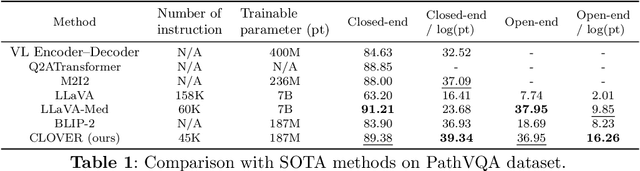
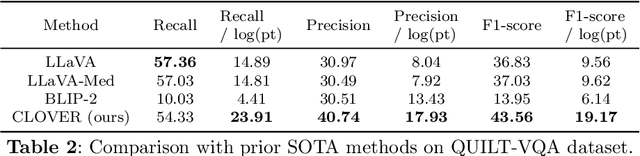

Abstract:The advent of vision-language models fosters the interactive conversations between AI-enabled models and humans. Yet applying these models into clinics must deal with daunting challenges around large-scale training data, financial, and computational resources. Here we propose a cost-effective instruction learning framework for conversational pathology named as CLOVER. CLOVER only trains a lightweight module and uses instruction tuning while freezing the parameters of the large language model. Instead of using costly GPT-4, we propose well-designed prompts on GPT-3.5 for building generation-based instructions, emphasizing the utility of pathological knowledge derived from the Internet source. To augment the use of instructions, we construct a high-quality set of template-based instructions in the context of digital pathology. From two benchmark datasets, our findings reveal the strength of hybrid-form instructions in the visual question-answer in pathology. Extensive results show the cost-effectiveness of CLOVER in answering both open-ended and closed-ended questions, where CLOVER outperforms strong baselines that possess 37 times more training parameters and use instruction data generated from GPT-4. Through the instruction tuning, CLOVER exhibits robustness of few-shot learning in the external clinical dataset. These findings demonstrate that cost-effective modeling of CLOVER could accelerate the adoption of rapid conversational applications in the landscape of digital pathology.
OpenMEDLab: An Open-source Platform for Multi-modality Foundation Models in Medicine
Mar 04, 2024



Abstract:The emerging trend of advancing generalist artificial intelligence, such as GPTv4 and Gemini, has reshaped the landscape of research (academia and industry) in machine learning and many other research areas. However, domain-specific applications of such foundation models (e.g., in medicine) remain untouched or often at their very early stages. It will require an individual set of transfer learning and model adaptation techniques by further expanding and injecting these models with domain knowledge and data. The development of such technologies could be largely accelerated if the bundle of data, algorithms, and pre-trained foundation models were gathered together and open-sourced in an organized manner. In this work, we present OpenMEDLab, an open-source platform for multi-modality foundation models. It encapsulates not only solutions of pioneering attempts in prompting and fine-tuning large language and vision models for frontline clinical and bioinformatic applications but also building domain-specific foundation models with large-scale multi-modal medical data. Importantly, it opens access to a group of pre-trained foundation models for various medical image modalities, clinical text, protein engineering, etc. Inspiring and competitive results are also demonstrated for each collected approach and model in a variety of benchmarks for downstream tasks. We welcome researchers in the field of medical artificial intelligence to continuously contribute cutting-edge methods and models to OpenMEDLab, which can be accessed via https://github.com/openmedlab.
AG-CRC: Anatomy-Guided Colorectal Cancer Segmentation in CT with Imperfect Anatomical Knowledge
Oct 07, 2023



Abstract:When delineating lesions from medical images, a human expert can always keep in mind the anatomical structure behind the voxels. However, although high-quality (though not perfect) anatomical information can be retrieved from computed tomography (CT) scans with modern deep learning algorithms, it is still an open problem how these automatically generated organ masks can assist in addressing challenging lesion segmentation tasks, such as the segmentation of colorectal cancer (CRC). In this paper, we develop a novel Anatomy-Guided segmentation framework to exploit the auto-generated organ masks to aid CRC segmentation from CT, namely AG-CRC. First, we obtain multi-organ segmentation (MOS) masks with existing MOS models (e.g., TotalSegmentor) and further derive a more robust organ of interest (OOI) mask that may cover most of the colon-rectum and CRC voxels. Then, we propose an anatomy-guided training patch sampling strategy by optimizing a heuristic gain function that considers both the proximity of important regions (e.g., the tumor or organs of interest) and sample diversity. Third, we design a novel self-supervised learning scheme inspired by the topology of tubular organs like the colon to boost the model performance further. Finally, we employ a masked loss scheme to guide the model to focus solely on the essential learning region. We extensively evaluate the proposed method on two CRC segmentation datasets, where substantial performance improvement (5% to 9% in Dice) is achieved over current state-of-the-art medical image segmentation models, and the ablation studies further evidence the efficacy of every proposed component.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge