Mianxin Liu
PathOrchestra: A Comprehensive Foundation Model for Computational Pathology with Over 100 Diverse Clinical-Grade Tasks
Mar 31, 2025Abstract:The complexity and variability inherent in high-resolution pathological images present significant challenges in computational pathology. While pathology foundation models leveraging AI have catalyzed transformative advancements, their development demands large-scale datasets, considerable storage capacity, and substantial computational resources. Furthermore, ensuring their clinical applicability and generalizability requires rigorous validation across a broad spectrum of clinical tasks. Here, we present PathOrchestra, a versatile pathology foundation model trained via self-supervised learning on a dataset comprising 300K pathological slides from 20 tissue and organ types across multiple centers. The model was rigorously evaluated on 112 clinical tasks using a combination of 61 private and 51 public datasets. These tasks encompass digital slide preprocessing, pan-cancer classification, lesion identification, multi-cancer subtype classification, biomarker assessment, gene expression prediction, and the generation of structured reports. PathOrchestra demonstrated exceptional performance across 27,755 WSIs and 9,415,729 ROIs, achieving over 0.950 accuracy in 47 tasks, including pan-cancer classification across various organs, lymphoma subtype diagnosis, and bladder cancer screening. Notably, it is the first model to generate structured reports for high-incidence colorectal cancer and diagnostically complex lymphoma-areas that are infrequently addressed by foundational models but hold immense clinical potential. Overall, PathOrchestra exemplifies the feasibility and efficacy of a large-scale, self-supervised pathology foundation model, validated across a broad range of clinical-grade tasks. Its high accuracy and reduced reliance on extensive data annotation underline its potential for clinical integration, offering a pathway toward more efficient and high-quality medical services.
End-to-end Triple-domain PET Enhancement: A Hybrid Denoising-and-reconstruction Framework for Reconstructing Standard-dose PET Images from Low-dose PET Sinograms
Dec 04, 2024
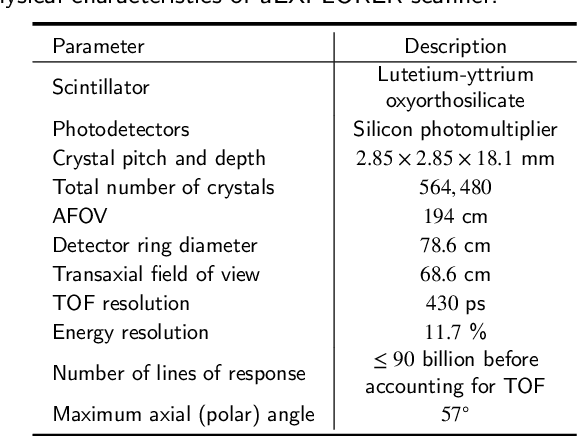

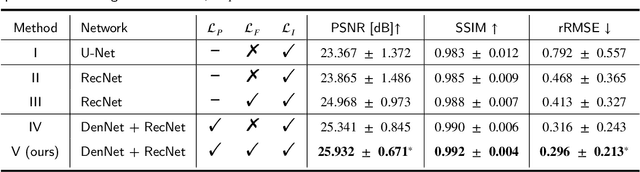
Abstract:As a sensitive functional imaging technique, positron emission tomography (PET) plays a critical role in early disease diagnosis. However, obtaining a high-quality PET image requires injecting a sufficient dose (standard dose) of radionuclides into the body, which inevitably poses radiation hazards to patients. To mitigate radiation hazards, the reconstruction of standard-dose PET (SPET) from low-dose PET (LPET) is desired. According to imaging theory, PET reconstruction process involves multiple domains (e.g., projection domain and image domain), and a significant portion of the difference between SPET and LPET arises from variations in the noise levels introduced during the sampling of raw data as sinograms. In light of these two facts, we propose an end-to-end TriPle-domain LPET EnhancemenT (TriPLET) framework, by leveraging the advantages of a hybrid denoising-and-reconstruction process and a triple-domain representation (i.e., sinograms, frequency spectrum maps, and images) to reconstruct SPET images from LPET sinograms. Specifically, TriPLET consists of three sequentially coupled components including 1) a Transformer-assisted denoising network that denoises the inputted LPET sinograms in the projection domain, 2) a discrete-wavelet-transform-based reconstruction network that further reconstructs SPET from LPET in the wavelet domain, and 3) a pair-based adversarial network that evaluates the reconstructed SPET images in the image domain. Extensive experiments on the real PET dataset demonstrate that our proposed TriPLET can reconstruct SPET images with the highest similarity and signal-to-noise ratio to real data, compared with state-of-the-art methods.
BrainMVP: Multi-modal Vision Pre-training for Brain Image Analysis using Multi-parametric MRI
Oct 14, 2024



Abstract:Accurate diagnosis of brain abnormalities is greatly enhanced by the inclusion of complementary multi-parametric MRI imaging data. There is significant potential to develop a universal pre-training model that can be quickly adapted for image modalities and various clinical scenarios. However, current models often rely on uni-modal image data, neglecting the cross-modal correlations among different image modalities or struggling to scale up pre-training in the presence of missing modality data. In this paper, we propose BrainMVP, a multi-modal vision pre-training framework for brain image analysis using multi-parametric MRI scans. First, we collect 16,022 brain MRI scans (over 2.4 million images), encompassing eight MRI modalities sourced from a diverse range of centers and devices. Then, a novel pre-training paradigm is proposed for the multi-modal MRI data, addressing the issue of missing modalities and achieving multi-modal information fusion. Cross-modal reconstruction is explored to learn distinctive brain image embeddings and efficient modality fusion capabilities. A modality-wise data distillation module is proposed to extract the essence representation of each MR image modality for both the pre-training and downstream application purposes. Furthermore, we introduce a modality-aware contrastive learning module to enhance the cross-modality association within a study. Extensive experiments on downstream tasks demonstrate superior performance compared to state-of-the-art pre-training methods in the medical domain, with Dice Score improvement of 0.28%-14.47% across six segmentation benchmarks and a consistent accuracy improvement of 0.65%-18.07% in four individual classification tasks.
CIResDiff: A Clinically-Informed Residual Diffusion Model for Predicting Idiopathic Pulmonary Fibrosis Progression
Aug 05, 2024


Abstract:The progression of Idiopathic Pulmonary Fibrosis (IPF) significantly correlates with higher patient mortality rates. Early detection of IPF progression is critical for initiating timely treatment, which can effectively slow down the advancement of the disease. However, the current clinical criteria define disease progression requiring two CT scans with a one-year interval, presenting a dilemma: a disease progression is identified only after the disease has already progressed. To this end, in this paper, we develop a novel diffusion model to accurately predict the progression of IPF by generating patient's follow-up CT scan from the initial CT scan. Specifically, from the clinical prior knowledge, we tailor improvements to the traditional diffusion model and propose a Clinically-Informed Residual Diffusion model, called CIResDiff. The key innovations of CIResDiff include 1) performing the target region pre-registration to align the lung regions of two CT scans at different time points for reducing the generation difficulty, 2) adopting the residual diffusion instead of traditional diffusion to enable the model focus more on differences (i.e., lesions) between the two CT scans rather than the largely identical anatomical content, and 3) designing the clinically-informed process based on CLIP technology to integrate lung function information which is highly relevant to diagnosis into the reverse process for assisting generation. Extensive experiments on clinical data demonstrate that our approach can outperform state-of-the-art methods and effectively predict the progression of IPF.
A dual-task mutual learning framework for predicting post-thrombectomy cerebral hemorrhage
Aug 01, 2024Abstract:Ischemic stroke is a severe condition caused by the blockage of brain blood vessels, and can lead to the death of brain tissue due to oxygen deprivation. Thrombectomy has become a common treatment choice for ischemic stroke due to its immediate effectiveness. But, it carries the risk of postoperative cerebral hemorrhage. Clinically, multiple CT scans within 0-72 hours post-surgery are used to monitor for hemorrhage. However, this approach exposes radiation dose to patients, and may delay the detection of cerebral hemorrhage. To address this dilemma, we propose a novel prediction framework for measuring postoperative cerebral hemorrhage using only the patient's initial CT scan. Specifically, we introduce a dual-task mutual learning framework to takes the initial CT scan as input and simultaneously estimates both the follow-up CT scan and prognostic label to predict the occurrence of postoperative cerebral hemorrhage. Our proposed framework incorporates two attention mechanisms, i.e., self-attention and interactive attention. Specifically, the self-attention mechanism allows the model to focus more on high-density areas in the image, which are critical for diagnosis (i.e., potential hemorrhage areas). The interactive attention mechanism further models the dependencies between the interrelated generation and classification tasks, enabling both tasks to perform better than the case when conducted individually. Validated on clinical data, our method can generate follow-up CT scans better than state-of-the-art methods, and achieves an accuracy of 86.37% in predicting follow-up prognostic labels. Thus, our work thus contributes to the timely screening of post-thrombectomy cerebral hemorrhage, and could significantly reform the clinical process of thrombectomy and other similar operations related to stroke.
A Progressive Single-Modality to Multi-Modality Classification Framework for Alzheimer's Disease Sub-type Diagnosis
Jul 26, 2024
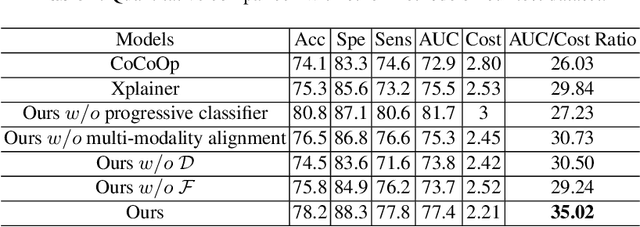
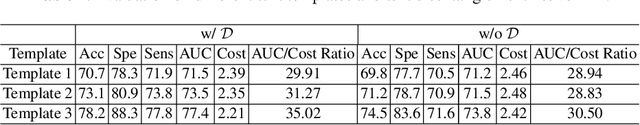
Abstract:The current clinical diagnosis framework of Alzheimer's disease (AD) involves multiple modalities acquired from multiple diagnosis stages, each with distinct usage and cost. Previous AD diagnosis research has predominantly focused on how to directly fuse multiple modalities for an end-to-end one-stage diagnosis, which practically requires a high cost in data acquisition. Moreover, a significant part of these methods diagnose AD without considering clinical guideline and cannot offer accurate sub-type diagnosis. In this paper, by exploring inter-correlation among multiple modalities, we propose a novel progressive AD sub-type diagnosis framework, aiming to give diagnosis results based on easier-to-access modalities in earlier low-cost stages, instead of modalities from all stages. Specifically, first, we design 1) a text disentanglement network for better processing tabular data collected in the initial stage, and 2) a modality fusion module for fusing multi-modality features separately. Second, we align features from modalities acquired in earlier low-cost stage(s) with later high-cost stage(s) to give accurate diagnosis without actual modality acquisition in later-stage(s) for saving cost. Furthermore, we follow the clinical guideline to align features at each stage for achieving sub-type diagnosis. Third, we leverage a progressive classifier that can progressively include additional acquired modalities (if needed) for diagnosis, to achieve the balance between diagnosis cost and diagnosis performance. We evaluate our proposed framework on large diverse public and in-home datasets (8280 in total) and achieve superior performance over state-of-the-art methods. Our codes will be released after the acceptance.
Cost-effective Instruction Learning for Pathology Vision and Language Analysis
Jul 25, 2024
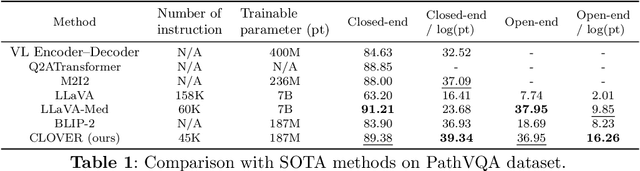
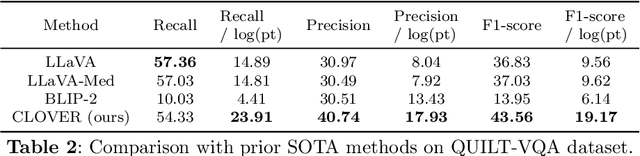

Abstract:The advent of vision-language models fosters the interactive conversations between AI-enabled models and humans. Yet applying these models into clinics must deal with daunting challenges around large-scale training data, financial, and computational resources. Here we propose a cost-effective instruction learning framework for conversational pathology named as CLOVER. CLOVER only trains a lightweight module and uses instruction tuning while freezing the parameters of the large language model. Instead of using costly GPT-4, we propose well-designed prompts on GPT-3.5 for building generation-based instructions, emphasizing the utility of pathological knowledge derived from the Internet source. To augment the use of instructions, we construct a high-quality set of template-based instructions in the context of digital pathology. From two benchmark datasets, our findings reveal the strength of hybrid-form instructions in the visual question-answer in pathology. Extensive results show the cost-effectiveness of CLOVER in answering both open-ended and closed-ended questions, where CLOVER outperforms strong baselines that possess 37 times more training parameters and use instruction data generated from GPT-4. Through the instruction tuning, CLOVER exhibits robustness of few-shot learning in the external clinical dataset. These findings demonstrate that cost-effective modeling of CLOVER could accelerate the adoption of rapid conversational applications in the landscape of digital pathology.
Mining fMRI Dynamics with Parcellation Prior for Brain Disease Diagnosis
May 04, 2023



Abstract:To characterize atypical brain dynamics under diseases, prevalent studies investigate functional magnetic resonance imaging (fMRI). However, most of the existing analyses compress rich spatial-temporal information as the brain functional networks (BFNs) and directly investigate the whole-brain network without neurological priors about functional subnetworks. We thus propose a novel graph learning framework to mine fMRI signals with topological priors from brain parcellation for disease diagnosis. Specifically, we 1) detect diagnosis-related temporal features using a "Transformer" for a higher-level BFN construction, and process it with a following graph convolutional network, and 2) apply an attention-based multiple instance learning strategy to emphasize the disease-affected subnetworks to further enhance the diagnosis performance and interpretability. Experiments demonstrate higher effectiveness of our method than compared methods in the diagnosis of early mild cognitive impairment. More importantly, our method is capable of localizing crucial brain subnetworks during the diagnosis, providing insights into the pathogenic source of mild cognitive impairment.
Structure-aware registration network for liver DCE-CT images
Mar 08, 2023



Abstract:Image registration of liver dynamic contrast-enhanced computed tomography (DCE-CT) is crucial for diagnosis and image-guided surgical planning of liver cancer. However, intensity variations due to the flow of contrast agents combined with complex spatial motion induced by respiration brings great challenge to existing intensity-based registration methods. To address these problems, we propose a novel structure-aware registration method by incorporating structural information of related organs with segmentation-guided deep registration network. Existing segmentation-guided registration methods only focus on volumetric registration inside the paired organ segmentations, ignoring the inherent attributes of their anatomical structures. In addition, such paired organ segmentations are not always available in DCE-CT images due to the flow of contrast agents. Different from existing segmentation-guided registration methods, our proposed method extracts structural information in hierarchical geometric perspectives of line and surface. Then, according to the extracted structural information, structure-aware constraints are constructed and imposed on the forward and backward deformation field simultaneously. In this way, all available organ segmentations, including unpaired ones, can be fully utilized to avoid the side effect of contrast agent and preserve the topology of organs during registration. Extensive experiments on an in-house liver DCE-CT dataset and a public LiTS dataset show that our proposed method can achieve higher registration accuracy and preserve anatomical structure more effectively than state-of-the-art methods.
Deep learning reveals the common spectrum underlying multiple brain disorders in youth and elders from brain functional networks
Feb 23, 2023



Abstract:Brain disorders in the early and late life of humans potentially share pathological alterations in brain functions. However, the key evidence from neuroimaging data for pathological commonness remains unrevealed. To explore this hypothesis, we build a deep learning model, using multi-site functional magnetic resonance imaging data (N=4,410, 6 sites), for classifying 5 different brain disorders from healthy controls, with a set of common features. Our model achieves 62.6(1.9)% overall classification accuracy on data from the 6 investigated sites and detects a set of commonly affected functional subnetworks at different spatial scales, including default mode, executive control, visual, and limbic networks. In the deep-layer feature representation for individual data, we observe young and aging patients with disorders are continuously distributed, which is in line with the clinical concept of the "spectrum of disorders". The revealed spectrum underlying early- and late-life brain disorders promotes the understanding of disorder comorbidities in the lifespan.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge