Zhongxiang Ding
Towards General Text-guided Image Synthesis for Customized Multimodal Brain MRI Generation
Sep 25, 2024



Abstract:Multimodal brain magnetic resonance (MR) imaging is indispensable in neuroscience and neurology. However, due to the accessibility of MRI scanners and their lengthy acquisition time, multimodal MR images are not commonly available. Current MR image synthesis approaches are typically trained on independent datasets for specific tasks, leading to suboptimal performance when applied to novel datasets and tasks. Here, we present TUMSyn, a Text-guided Universal MR image Synthesis generalist model, which can flexibly generate brain MR images with demanded imaging metadata from routinely acquired scans guided by text prompts. To ensure TUMSyn's image synthesis precision, versatility, and generalizability, we first construct a brain MR database comprising 31,407 3D images with 7 MRI modalities from 13 centers. We then pre-train an MRI-specific text encoder using contrastive learning to effectively control MR image synthesis based on text prompts. Extensive experiments on diverse datasets and physician assessments indicate that TUMSyn can generate clinically meaningful MR images with specified imaging metadata in supervised and zero-shot scenarios. Therefore, TUMSyn can be utilized along with acquired MR scan(s) to facilitate large-scale MRI-based screening and diagnosis of brain diseases.
Prototype Learning Guided Hybrid Network for Breast Tumor Segmentation in DCE-MRI
Aug 11, 2024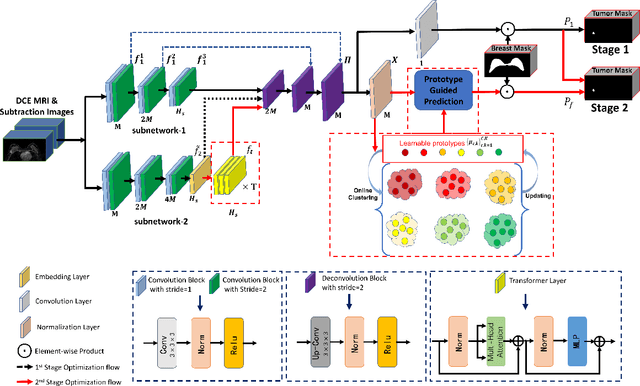
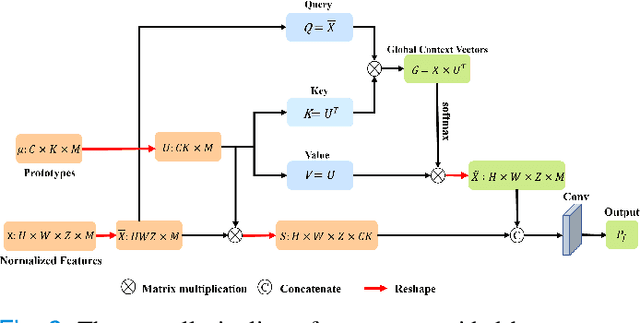
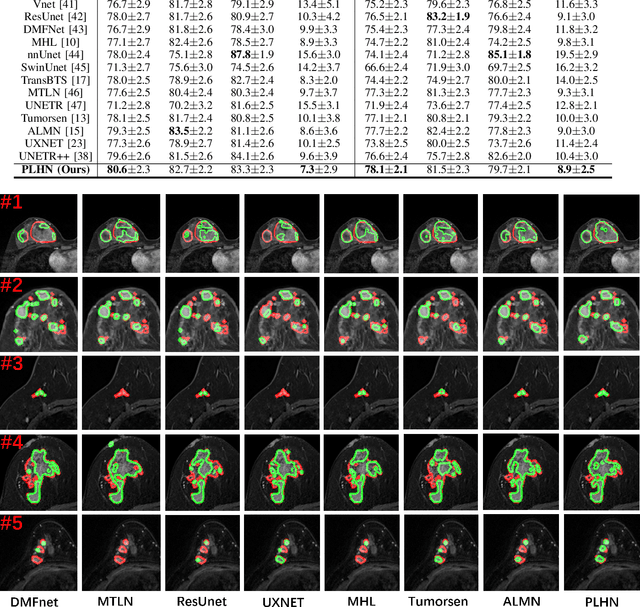
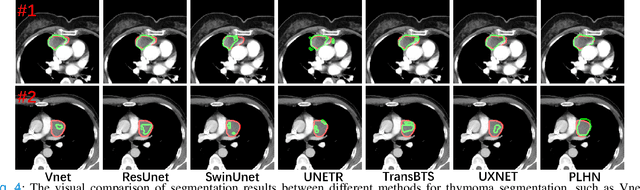
Abstract:Automated breast tumor segmentation on the basis of dynamic contrast-enhancement magnetic resonance imaging (DCE-MRI) has shown great promise in clinical practice, particularly for identifying the presence of breast disease. However, accurate segmentation of breast tumor is a challenging task, often necessitating the development of complex networks. To strike an optimal trade-off between computational costs and segmentation performance, we propose a hybrid network via the combination of convolution neural network (CNN) and transformer layers. Specifically, the hybrid network consists of a encoder-decoder architecture by stacking convolution and decovolution layers. Effective 3D transformer layers are then implemented after the encoder subnetworks, to capture global dependencies between the bottleneck features. To improve the efficiency of hybrid network, two parallel encoder subnetworks are designed for the decoder and the transformer layers, respectively. To further enhance the discriminative capability of hybrid network, a prototype learning guided prediction module is proposed, where the category-specified prototypical features are calculated through on-line clustering. All learned prototypical features are finally combined with the features from decoder for tumor mask prediction. The experimental results on private and public DCE-MRI datasets demonstrate that the proposed hybrid network achieves superior performance than the state-of-the-art (SOTA) methods, while maintaining balance between segmentation accuracy and computation cost. Moreover, we demonstrate that automatically generated tumor masks can be effectively applied to identify HER2-positive subtype from HER2-negative subtype with the similar accuracy to the analysis based on manual tumor segmentation. The source code is available at https://github.com/ZhouL-lab/PLHN.
A dual-task mutual learning framework for predicting post-thrombectomy cerebral hemorrhage
Aug 01, 2024Abstract:Ischemic stroke is a severe condition caused by the blockage of brain blood vessels, and can lead to the death of brain tissue due to oxygen deprivation. Thrombectomy has become a common treatment choice for ischemic stroke due to its immediate effectiveness. But, it carries the risk of postoperative cerebral hemorrhage. Clinically, multiple CT scans within 0-72 hours post-surgery are used to monitor for hemorrhage. However, this approach exposes radiation dose to patients, and may delay the detection of cerebral hemorrhage. To address this dilemma, we propose a novel prediction framework for measuring postoperative cerebral hemorrhage using only the patient's initial CT scan. Specifically, we introduce a dual-task mutual learning framework to takes the initial CT scan as input and simultaneously estimates both the follow-up CT scan and prognostic label to predict the occurrence of postoperative cerebral hemorrhage. Our proposed framework incorporates two attention mechanisms, i.e., self-attention and interactive attention. Specifically, the self-attention mechanism allows the model to focus more on high-density areas in the image, which are critical for diagnosis (i.e., potential hemorrhage areas). The interactive attention mechanism further models the dependencies between the interrelated generation and classification tasks, enabling both tasks to perform better than the case when conducted individually. Validated on clinical data, our method can generate follow-up CT scans better than state-of-the-art methods, and achieves an accuracy of 86.37% in predicting follow-up prognostic labels. Thus, our work thus contributes to the timely screening of post-thrombectomy cerebral hemorrhage, and could significantly reform the clinical process of thrombectomy and other similar operations related to stroke.
Accurate 3D Prediction of Missing Teeth in Diverse Patterns for Precise Dental Implant Planning
Jul 16, 2023



Abstract:In recent years, the demand for dental implants has surged, driven by their high success rates and esthetic advantages. However, accurate prediction of missing teeth for precise digital implant planning remains a challenge due to the intricate nature of dental structures and the variability in tooth loss patterns. This study presents a novel framework for accurate prediction of missing teeth in different patterns, facilitating digital implant planning. The proposed framework begins by estimating point-to-point correspondence among a dataset of dental mesh models reconstructed from CBCT images of healthy subjects. Subsequently, tooth dictionaries are constructed for each tooth type, encoding their position and shape information based on the established point-to-point correspondence. To predict missing teeth in a given dental mesh model, sparse coefficients are learned by sparsely representing adjacent teeth of the missing teeth using the corresponding tooth dictionaries. These coefficients are then applied to the dictionaries of the missing teeth to generate accurate predictions of their positions and shapes. The evaluation results on real subjects shows that our proposed framework achieves an average prediction error of 1.04mm for predictions of single missing tooth and an average prediction error of 1.33mm for the prediction of 14 missing teeth, which demonstrates its capability of accurately predicting missing teeth in various patterns. By accurately predicting missing teeth, dental professionals can improve the planning and placement of dental implants, leading to better esthetic and functional outcomes for patients undergoing dental implant procedures.
Construction of unbiased dental template and parametric dental model for precision digital dentistry
Apr 07, 2023Abstract:Dental template and parametric dental models are important tools for various applications in digital dentistry. However, constructing an unbiased dental template and accurate parametric dental models remains a challenging task due to the complex anatomical and morphological dental structures and also low volume ratio of the teeth. In this study, we develop an unbiased dental template by constructing an accurate dental atlas from CBCT images with guidance of teeth segmentation. First, to address the challenges, we propose to enhance the CBCT images and their segmentation images, including image cropping, image masking and segmentation intensity reassigning. Then, we further use the segmentation images to perform co-registration with the CBCT images to generate an accurate dental atlas, from which an unbiased dental template can be generated. By leveraging the unbiased dental template, we construct parametric dental models by estimating point-to-point correspondences between the dental models and employing Principal Component Analysis to determine shape subspaces of the parametric dental models. A total of 159 CBCT images of real subjects are collected to perform the constructions. Experimental results demonstrate effectiveness of our proposed method in constructing unbiased dental template and parametric dental model. The developed dental template and parametric dental models are available at https://github.com/Marvin0724/Teeth_template.
Cross-Site Severity Assessment of COVID-19 from CT Images via Domain Adaptation
Sep 08, 2021



Abstract:Early and accurate severity assessment of Coronavirus disease 2019 (COVID-19) based on computed tomography (CT) images offers a great help to the estimation of intensive care unit event and the clinical decision of treatment planning. To augment the labeled data and improve the generalization ability of the classification model, it is necessary to aggregate data from multiple sites. This task faces several challenges including class imbalance between mild and severe infections, domain distribution discrepancy between sites, and presence of heterogeneous features. In this paper, we propose a novel domain adaptation (DA) method with two components to address these problems. The first component is a stochastic class-balanced boosting sampling strategy that overcomes the imbalanced learning problem and improves the classification performance on poorly-predicted classes. The second component is a representation learning that guarantees three properties: 1) domain-transferability by prototype triplet loss, 2) discriminant by conditional maximum mean discrepancy loss, and 3) completeness by multi-view reconstruction loss. Particularly, we propose a domain translator and align the heterogeneous data to the estimated class prototypes (i.e., class centers) in a hyper-sphere manifold. Experiments on cross-site severity assessment of COVID-19 from CT images show that the proposed method can effectively tackle the imbalanced learning problem and outperform recent DA approaches.
A novel multiple instance learning framework for COVID-19 severity assessment via data augmentation and self-supervised learning
Feb 07, 2021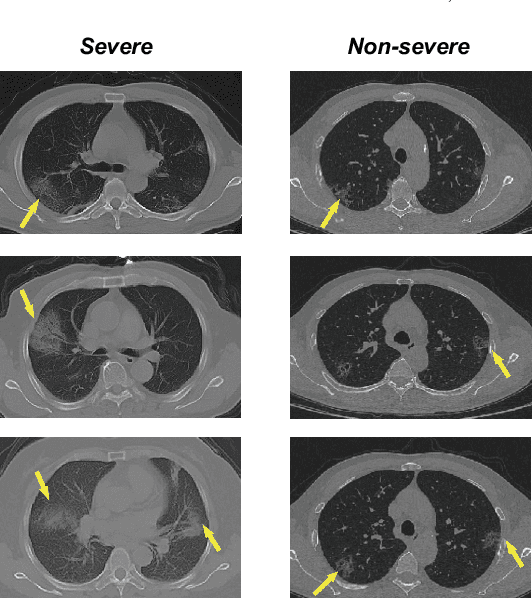
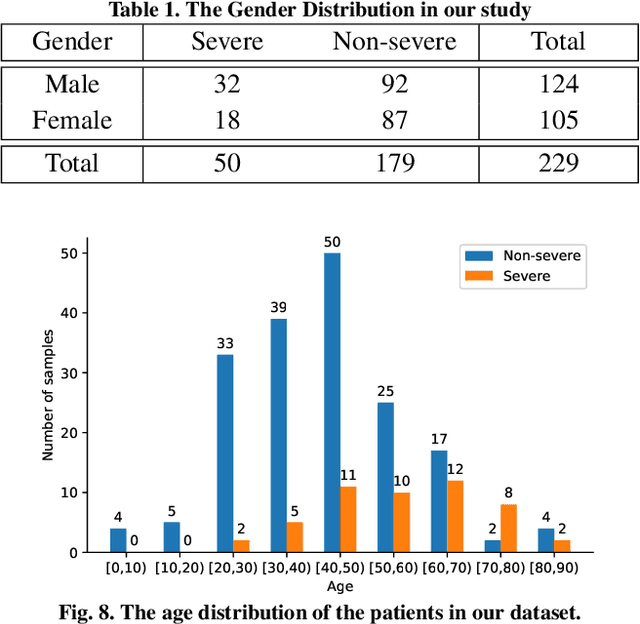
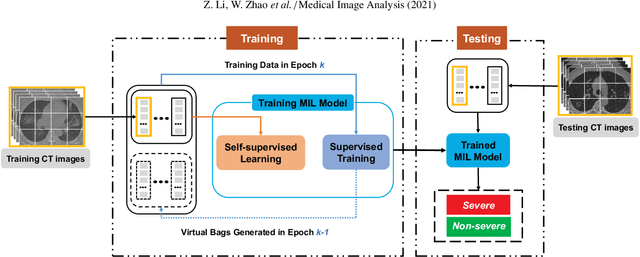
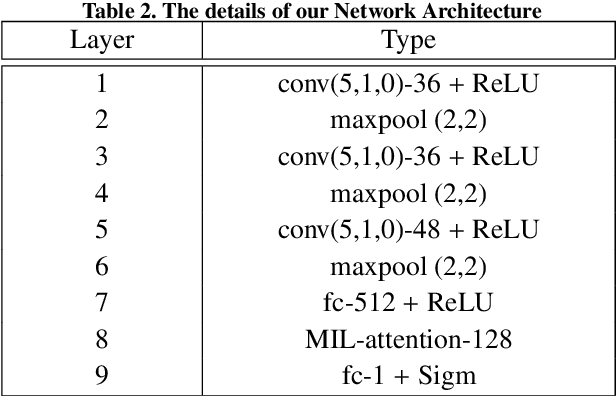
Abstract:How to fast and accurately assess the severity level of COVID-19 is an essential problem, when millions of people are suffering from the pandemic around the world. Currently, the chest CT is regarded as a popular and informative imaging tool for COVID-19 diagnosis. However, we observe that there are two issues -- weak annotation and insufficient data that may obstruct automatic COVID-19 severity assessment with CT images. To address these challenges, we propose a novel three-component method, i.e., 1) a deep multiple instance learning component with instance-level attention to jointly classify the bag and also weigh the instances, 2) a bag-level data augmentation component to generate virtual bags by reorganizing high confidential instances, and 3) a self-supervised pretext component to aid the learning process. We have systematically evaluated our method on the CT images of 229 COVID-19 cases, including 50 severe and 179 non-severe cases. Our method could obtain an average accuracy of 95.8%, with 93.6% sensitivity and 96.4% specificity, which outperformed previous works.
Dual-Sampling Attention Network for Diagnosis of COVID-19 from Community Acquired Pneumonia
May 20, 2020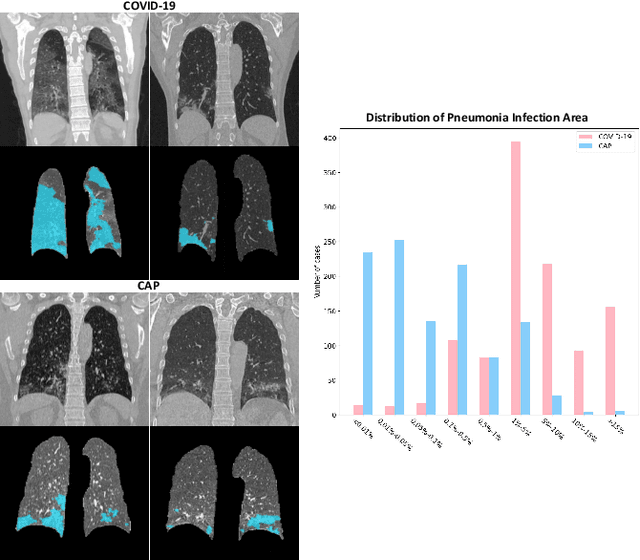
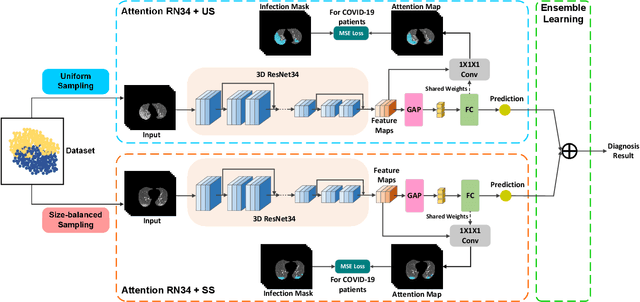
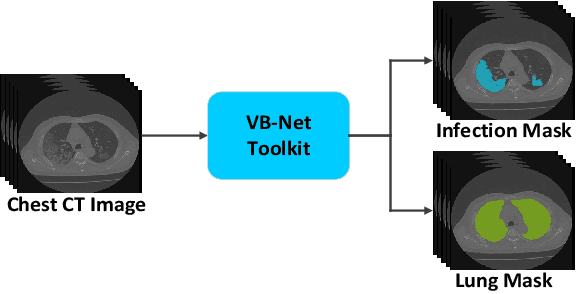
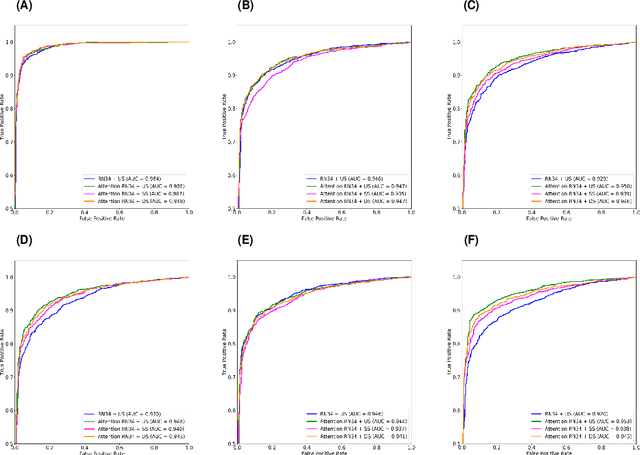
Abstract:The coronavirus disease (COVID-19) is rapidly spreading all over the world, and has infected more than 1,436,000 people in more than 200 countries and territories as of April 9, 2020. Detecting COVID-19 at early stage is essential to deliver proper healthcare to the patients and also to protect the uninfected population. To this end, we develop a dual-sampling attention network to automatically diagnose COVID- 19 from the community acquired pneumonia (CAP) in chest computed tomography (CT). In particular, we propose a novel online attention module with a 3D convolutional network (CNN) to focus on the infection regions in lungs when making decisions of diagnoses. Note that there exists imbalanced distribution of the sizes of the infection regions between COVID-19 and CAP, partially due to fast progress of COVID-19 after symptom onset. Therefore, we develop a dual-sampling strategy to mitigate the imbalanced learning. Our method is evaluated (to our best knowledge) upon the largest multi-center CT data for COVID-19 from 8 hospitals. In the training-validation stage, we collect 2186 CT scans from 1588 patients for a 5-fold cross-validation. In the testing stage, we employ another independent large-scale testing dataset including 2796 CT scans from 2057 patients. Results show that our algorithm can identify the COVID-19 images with the area under the receiver operating characteristic curve (AUC) value of 0.944, accuracy of 87.5%, sensitivity of 86.9%, specificity of 90.1%, and F1-score of 82.0%. With this performance, the proposed algorithm could potentially aid radiologists with COVID-19 diagnosis from CAP, especially in the early stage of the COVID-19 outbreak.
Hypergraph Learning for Identification of COVID-19 with CT Imaging
May 07, 2020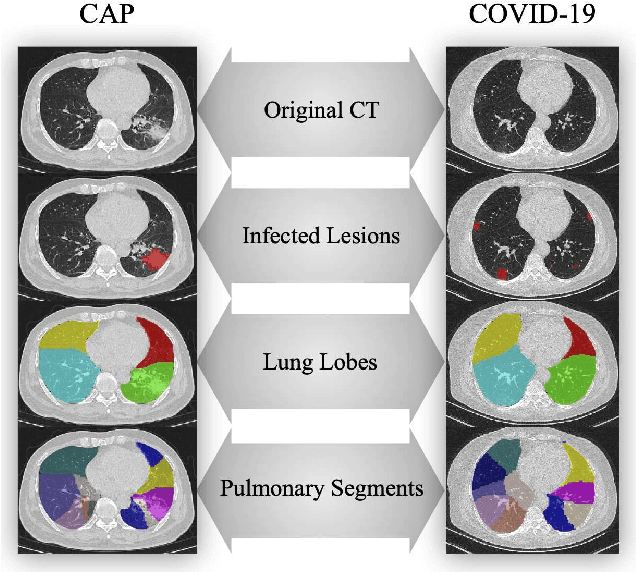
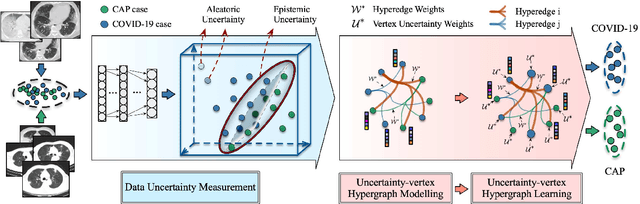
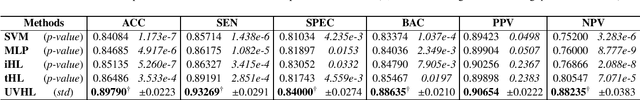
Abstract:The coronavirus disease, named COVID-19, has become the largest global public health crisis since it started in early 2020. CT imaging has been used as a complementary tool to assist early screening, especially for the rapid identification of COVID-19 cases from community acquired pneumonia (CAP) cases. The main challenge in early screening is how to model the confusing cases in the COVID-19 and CAP groups, with very similar clinical manifestations and imaging features. To tackle this challenge, we propose an Uncertainty Vertex-weighted Hypergraph Learning (UVHL) method to identify COVID-19 from CAP using CT images. In particular, multiple types of features (including regional features and radiomics features) are first extracted from CT image for each case. Then, the relationship among different cases is formulated by a hypergraph structure, with each case represented as a vertex in the hypergraph. The uncertainty of each vertex is further computed with an uncertainty score measurement and used as a weight in the hypergraph. Finally, a learning process of the vertex-weighted hypergraph is used to predict whether a new testing case belongs to COVID-19 or not. Experiments on a large multi-center pneumonia dataset, consisting of 2,148 COVID-19 cases and 1,182 CAP cases from five hospitals, are conducted to evaluate the performance of the proposed method. Results demonstrate the effectiveness and robustness of our proposed method on the identification of COVID-19 in comparison to state-of-the-art methods.
Adaptive Feature Selection Guided Deep Forest for COVID-19 Classification with Chest CT
May 07, 2020



Abstract:Chest computed tomography (CT) becomes an effective tool to assist the diagnosis of coronavirus disease-19 (COVID-19). Due to the outbreak of COVID-19 worldwide, using the computed-aided diagnosis technique for COVID-19 classification based on CT images could largely alleviate the burden of clinicians. In this paper, we propose an Adaptive Feature Selection guided Deep Forest (AFS-DF) for COVID-19 classification based on chest CT images. Specifically, we first extract location-specific features from CT images. Then, in order to capture the high-level representation of these features with the relatively small-scale data, we leverage a deep forest model to learn high-level representation of the features. Moreover, we propose a feature selection method based on the trained deep forest model to reduce the redundancy of features, where the feature selection could be adaptively incorporated with the COVID-19 classification model. We evaluated our proposed AFS-DF on COVID-19 dataset with 1495 patients of COVID-19 and 1027 patients of community acquired pneumonia (CAP). The accuracy (ACC), sensitivity (SEN), specificity (SPE) and AUC achieved by our method are 91.79%, 93.05%, 89.95% and 96.35%, respectively. Experimental results on the COVID-19 dataset suggest that the proposed AFS-DF achieves superior performance in COVID-19 vs. CAP classification, compared with 4 widely used machine learning methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge