Shengrui Li
Decouple Searching from Training: Scaling Data Mixing via Model Merging for Large Language Model Pre-training
Jan 31, 2026Abstract:Determining an effective data mixture is a key factor in Large Language Model (LLM) pre-training, where models must balance general competence with proficiency on hard tasks such as math and code. However, identifying an optimal mixture remains an open challenge, as existing approaches either rely on unreliable tiny-scale proxy experiments or require prohibitively expensive large-scale exploration. To address this, we propose Decouple Searching from Training Mix (DeMix), a novel framework that leverages model merging to predict optimal data ratios. Instead of training proxy models for every sampled mixture, DeMix trains component models on candidate datasets at scale and derives data mixture proxies via weighted model merging. This paradigm decouples search from training costs, enabling evaluation of unlimited sampled mixtures without extra training burden and thus facilitating better mixture discovery through more search trials. Extensive experiments demonstrate that DeMix breaks the trade-off between sufficiency, accuracy and efficiency, obtaining the optimal mixture with higher benchmark performance at lower search cost. Additionally, we release the DeMix Corpora, a comprehensive 22T-token dataset comprising high-quality pre-training data with validated mixtures to facilitate open research. Our code and DeMix Corpora is available at https://github.com/Lucius-lsr/DeMix.
NutePrune: Efficient Progressive Pruning with Numerous Teachers for Large Language Models
Feb 15, 2024



Abstract:The considerable size of Large Language Models (LLMs) presents notable deployment challenges, particularly on resource-constrained hardware. Structured pruning, offers an effective means to compress LLMs, thereby reducing storage costs and enhancing inference speed for more efficient utilization. In this work, we study data-efficient and resource-efficient structure pruning methods to obtain smaller yet still powerful models. Knowledge Distillation is well-suited for pruning, as the intact model can serve as an excellent teacher for pruned students. However, it becomes challenging in the context of LLMs due to memory constraints. To address this, we propose an efficient progressive Numerous-teacher pruning method (NutePrune). NutePrune mitigates excessive memory costs by loading only one intact model and integrating it with various masks and LoRA modules, enabling it to seamlessly switch between teacher and student roles. This approach allows us to leverage numerous teachers with varying capacities to progressively guide the pruned model, enhancing overall performance. Extensive experiments across various tasks demonstrate the effectiveness of NutePrune. In LLaMA-7B zero-shot experiments, NutePrune retains 97.17% of the performance of the original model at 20% sparsity and 95.07% at 25% sparsity.
AdapterGNN: Efficient Delta Tuning Improves Generalization Ability in Graph Neural Networks
Apr 19, 2023



Abstract:Fine-tuning pre-trained models has recently yielded remarkable performance gains in graph neural networks (GNNs). In addition to pre-training techniques, inspired by the latest work in the natural language fields, more recent work has shifted towards applying effective fine-tuning approaches, such as parameter-efficient tuning (delta tuning). However, given the substantial differences between GNNs and transformer-based models, applying such approaches directly to GNNs proved to be less effective. In this paper, we present a comprehensive comparison of delta tuning techniques for GNNs and propose a novel delta tuning method specifically designed for GNNs, called AdapterGNN. AdapterGNN preserves the knowledge of the large pre-trained model and leverages highly expressive adapters for GNNs, which can adapt to downstream tasks effectively with only a few parameters, while also improving the model's generalization ability on the downstream tasks. Extensive experiments show that AdapterGNN achieves higher evaluation performance (outperforming full fine-tuning by 1.4% and 5.5% in the chemistry and biology domains respectively, with only 5% of its parameters tuned) and lower generalization gaps compared to full fine-tuning. Moreover, we empirically show that a larger GNN model can have a worse generalization ability, which differs from the trend observed in large language models. We have also provided a theoretical justification for delta tuning can improve the generalization ability of GNNs by applying generalization bounds.
Hypergraph Learning for Identification of COVID-19 with CT Imaging
May 07, 2020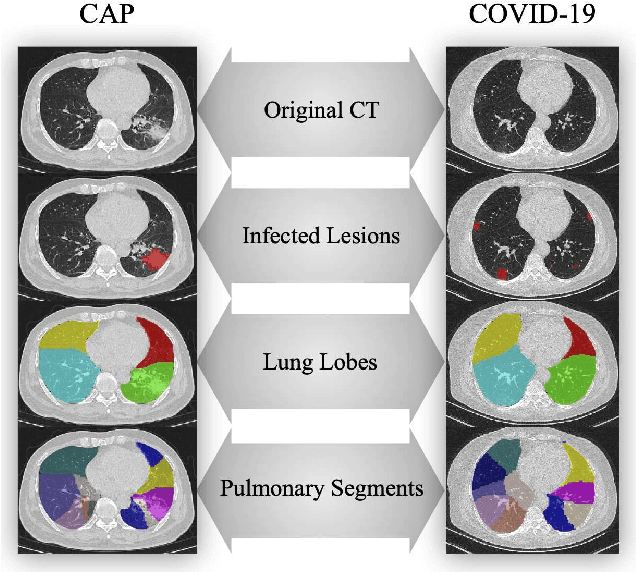
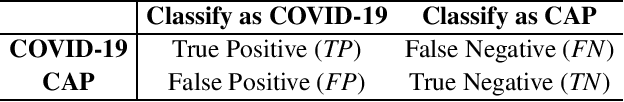
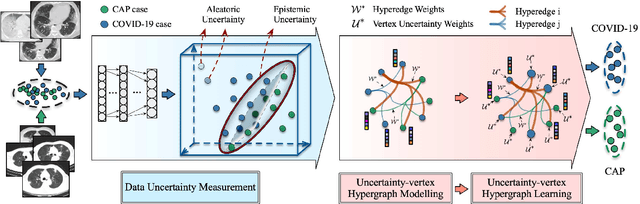
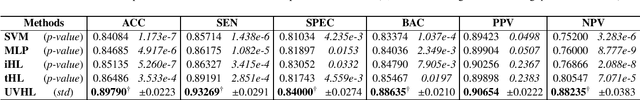
Abstract:The coronavirus disease, named COVID-19, has become the largest global public health crisis since it started in early 2020. CT imaging has been used as a complementary tool to assist early screening, especially for the rapid identification of COVID-19 cases from community acquired pneumonia (CAP) cases. The main challenge in early screening is how to model the confusing cases in the COVID-19 and CAP groups, with very similar clinical manifestations and imaging features. To tackle this challenge, we propose an Uncertainty Vertex-weighted Hypergraph Learning (UVHL) method to identify COVID-19 from CAP using CT images. In particular, multiple types of features (including regional features and radiomics features) are first extracted from CT image for each case. Then, the relationship among different cases is formulated by a hypergraph structure, with each case represented as a vertex in the hypergraph. The uncertainty of each vertex is further computed with an uncertainty score measurement and used as a weight in the hypergraph. Finally, a learning process of the vertex-weighted hypergraph is used to predict whether a new testing case belongs to COVID-19 or not. Experiments on a large multi-center pneumonia dataset, consisting of 2,148 COVID-19 cases and 1,182 CAP cases from five hospitals, are conducted to evaluate the performance of the proposed method. Results demonstrate the effectiveness and robustness of our proposed method on the identification of COVID-19 in comparison to state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge