Liming Xia
Deep-learning-based groupwise registration for motion correction of cardiac $T_1$ mapping
Jun 21, 2024Abstract:Quantitative $T_1$ mapping by MRI is an increasingly important tool for clinical assessment of cardiovascular diseases. The cardiac $T_1$ map is derived by fitting a known signal model to a series of baseline images, while the quality of this map can be deteriorated by involuntary respiratory and cardiac motion. To correct motion, a template image is often needed to register all baseline images, but the choice of template is nontrivial, leading to inconsistent performance sensitive to image contrast. In this work, we propose a novel deep-learning-based groupwise registration framework, which omits the need for a template, and registers all baseline images simultaneously. We design two groupwise losses for this registration framework: the first is a linear principal component analysis (PCA) loss that enforces alignment of baseline images irrespective of the intensity variation, and the second is an auxiliary relaxometry loss that enforces adherence of intensity profile to the signal model. We extensively evaluated our method, termed ``PCA-Relax'', and other baseline methods on an in-house cardiac MRI dataset including both pre- and post-contrast $T_1$ sequences. All methods were evaluated under three distinct training-and-evaluation strategies, namely, standard, one-shot, and test-time-adaptation. The proposed PCA-Relax showed further improved performance of registration and mapping over well-established baselines. The proposed groupwise framework is generic and can be adapted to applications involving multiple images.
PCA-Relax: Deep-learning-based groupwise registration for motion correction of cardiac $T_1$ mapping
Jun 18, 2024Abstract:Quantitative MRI (qMRI) is an increasingly important tool for clinical assessment of cardiovascular diseases. Quantitative maps are derived by fitting a known signal model to a series of baseline images, while the quality of the map can be deteriorated by involuntary respiratory and cardiac motion. To correct motion, a template image is often needed to register all baseline images, but the choice of template is nontrivial, leading to inconsistent performance sensitive to image contrast. In this work, we propose a novel deep-learning-based groupwise registration framework, which omits the need for a template, and registers all baseline images simultaneously. We design two groupwise losses for this registration framework: the first is a linear principal component analysis (PCA) loss that enforces alignment of baseline images irrespective of the intensity variation, and the second is an auxiliary relaxometry loss that enforces adherence of intensity profile to the signal model. We extensively evaluated our method, termed ``PCA-Relax'', and other baseline methods on an in-house cardiac MRI dataset including both pre- and post-contrast $T_1$ sequences. All methods were evaluated under three distinct training-and-evaluation strategies, namely, standard, one-shot, and test-time-adaptation. The proposed PCA-Relax showed further improved performance of registration and mapping over well-established baselines. The proposed groupwise framework is generic and can be adapted to applications involving multiple images.
Dual-Sampling Attention Network for Diagnosis of COVID-19 from Community Acquired Pneumonia
May 20, 2020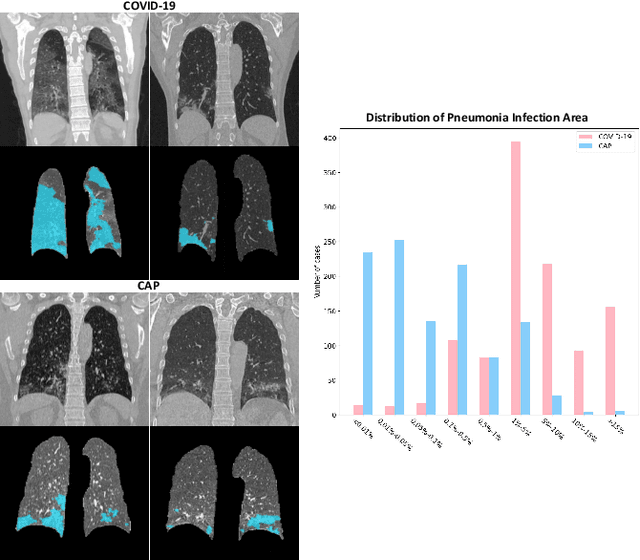
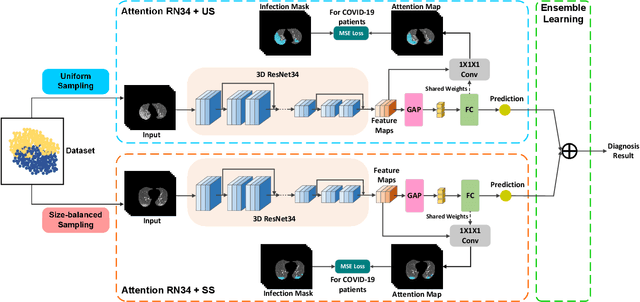
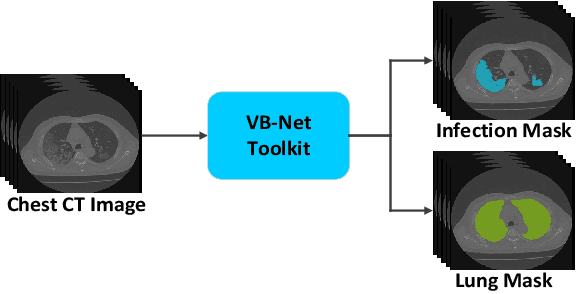
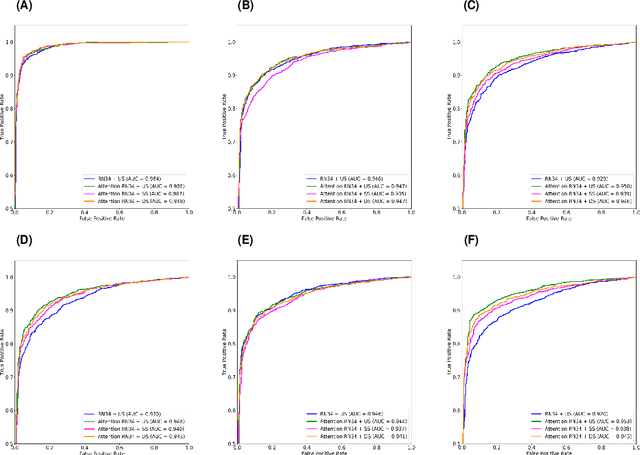
Abstract:The coronavirus disease (COVID-19) is rapidly spreading all over the world, and has infected more than 1,436,000 people in more than 200 countries and territories as of April 9, 2020. Detecting COVID-19 at early stage is essential to deliver proper healthcare to the patients and also to protect the uninfected population. To this end, we develop a dual-sampling attention network to automatically diagnose COVID- 19 from the community acquired pneumonia (CAP) in chest computed tomography (CT). In particular, we propose a novel online attention module with a 3D convolutional network (CNN) to focus on the infection regions in lungs when making decisions of diagnoses. Note that there exists imbalanced distribution of the sizes of the infection regions between COVID-19 and CAP, partially due to fast progress of COVID-19 after symptom onset. Therefore, we develop a dual-sampling strategy to mitigate the imbalanced learning. Our method is evaluated (to our best knowledge) upon the largest multi-center CT data for COVID-19 from 8 hospitals. In the training-validation stage, we collect 2186 CT scans from 1588 patients for a 5-fold cross-validation. In the testing stage, we employ another independent large-scale testing dataset including 2796 CT scans from 2057 patients. Results show that our algorithm can identify the COVID-19 images with the area under the receiver operating characteristic curve (AUC) value of 0.944, accuracy of 87.5%, sensitivity of 86.9%, specificity of 90.1%, and F1-score of 82.0%. With this performance, the proposed algorithm could potentially aid radiologists with COVID-19 diagnosis from CAP, especially in the early stage of the COVID-19 outbreak.
Hypergraph Learning for Identification of COVID-19 with CT Imaging
May 07, 2020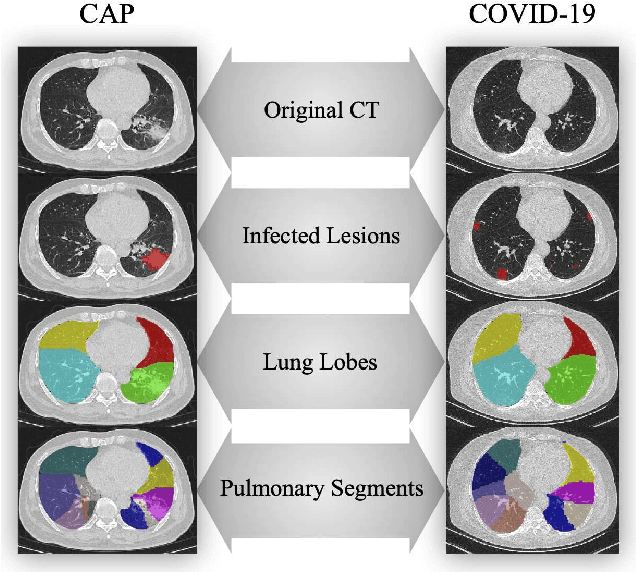
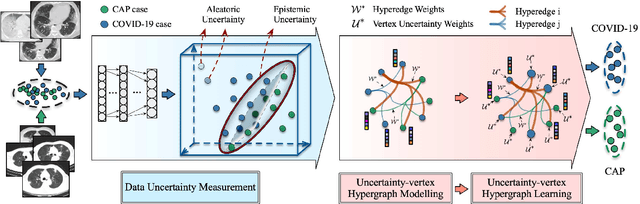
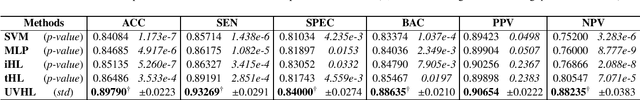
Abstract:The coronavirus disease, named COVID-19, has become the largest global public health crisis since it started in early 2020. CT imaging has been used as a complementary tool to assist early screening, especially for the rapid identification of COVID-19 cases from community acquired pneumonia (CAP) cases. The main challenge in early screening is how to model the confusing cases in the COVID-19 and CAP groups, with very similar clinical manifestations and imaging features. To tackle this challenge, we propose an Uncertainty Vertex-weighted Hypergraph Learning (UVHL) method to identify COVID-19 from CAP using CT images. In particular, multiple types of features (including regional features and radiomics features) are first extracted from CT image for each case. Then, the relationship among different cases is formulated by a hypergraph structure, with each case represented as a vertex in the hypergraph. The uncertainty of each vertex is further computed with an uncertainty score measurement and used as a weight in the hypergraph. Finally, a learning process of the vertex-weighted hypergraph is used to predict whether a new testing case belongs to COVID-19 or not. Experiments on a large multi-center pneumonia dataset, consisting of 2,148 COVID-19 cases and 1,182 CAP cases from five hospitals, are conducted to evaluate the performance of the proposed method. Results demonstrate the effectiveness and robustness of our proposed method on the identification of COVID-19 in comparison to state-of-the-art methods.
Adaptive Feature Selection Guided Deep Forest for COVID-19 Classification with Chest CT
May 07, 2020



Abstract:Chest computed tomography (CT) becomes an effective tool to assist the diagnosis of coronavirus disease-19 (COVID-19). Due to the outbreak of COVID-19 worldwide, using the computed-aided diagnosis technique for COVID-19 classification based on CT images could largely alleviate the burden of clinicians. In this paper, we propose an Adaptive Feature Selection guided Deep Forest (AFS-DF) for COVID-19 classification based on chest CT images. Specifically, we first extract location-specific features from CT images. Then, in order to capture the high-level representation of these features with the relatively small-scale data, we leverage a deep forest model to learn high-level representation of the features. Moreover, we propose a feature selection method based on the trained deep forest model to reduce the redundancy of features, where the feature selection could be adaptively incorporated with the COVID-19 classification model. We evaluated our proposed AFS-DF on COVID-19 dataset with 1495 patients of COVID-19 and 1027 patients of community acquired pneumonia (CAP). The accuracy (ACC), sensitivity (SEN), specificity (SPE) and AUC achieved by our method are 91.79%, 93.05%, 89.95% and 96.35%, respectively. Experimental results on the COVID-19 dataset suggest that the proposed AFS-DF achieves superior performance in COVID-19 vs. CAP classification, compared with 4 widely used machine learning methods.
Diagnosis of Coronavirus Disease 2019 (COVID-19) with Structured Latent Multi-View Representation Learning
May 06, 2020



Abstract:Recently, the outbreak of Coronavirus Disease 2019 (COVID-19) has spread rapidly across the world. Due to the large number of affected patients and heavy labor for doctors, computer-aided diagnosis with machine learning algorithm is urgently needed, and could largely reduce the efforts of clinicians and accelerate the diagnosis process. Chest computed tomography (CT) has been recognized as an informative tool for diagnosis of the disease. In this study, we propose to conduct the diagnosis of COVID-19 with a series of features extracted from CT images. To fully explore multiple features describing CT images from different views, a unified latent representation is learned which can completely encode information from different aspects of features and is endowed with promising class structure for separability. Specifically, the completeness is guaranteed with a group of backward neural networks (each for one type of features), while by using class labels the representation is enforced to be compact within COVID-19/community-acquired pneumonia (CAP) and also a large margin is guaranteed between different types of pneumonia. In this way, our model can well avoid overfitting compared to the case of directly projecting highdimensional features into classes. Extensive experimental results show that the proposed method outperforms all comparison methods, and rather stable performances are observed when varying the numbers of training data.
Large-Scale Screening of COVID-19 from Community Acquired Pneumonia using Infection Size-Aware Classification
Mar 22, 2020



Abstract:The worldwide spread of coronavirus disease (COVID-19) has become a threatening risk for global public health. It is of great importance to rapidly and accurately screen patients with COVID-19 from community acquired pneumonia (CAP). In this study, a total of 1658 patients with COVID-19 and 1027 patients of CAP underwent thin-section CT. All images were preprocessed to obtain the segmentations of both infections and lung fields, which were used to extract location-specific features. An infection Size Aware Random Forest method (iSARF) was proposed, in which subjects were automated categorized into groups with different ranges of infected lesion sizes, followed by random forests in each group for classification. Experimental results show that the proposed method yielded sensitivity of 0.907, specificity of 0.833, and accuracy of 0.879 under five-fold cross-validation. Large performance margins against comparison methods were achieved especially for the cases with infection size in the medium range, from 0.01% to 10%. The further inclusion of Radiomics features show slightly improvement. It is anticipated that our proposed framework could assist clinical decision making.
The Domain Shift Problem of Medical Image Segmentation and Vendor-Adaptation by Unet-GAN
Oct 30, 2019



Abstract:Convolutional neural network (CNN), in particular the Unet, is a powerful method for medical image segmentation. To date Unet has demonstrated state-of-art performance in many complex medical image segmentation tasks, especially under the condition when the training and testing data share the same distribution (i.e. come from the same source domain). However, in clinical practice, medical images are acquired from different vendors and centers. The performance of a U-Net trained from a particular source domain, when transferred to a different target domain (e.g. different vendor, acquisition parameter), can drop unexpectedly. Collecting a large amount of annotation from each new domain to retrain the U-Net is expensive, tedious, and practically impossible. In this work, we proposed a generic framework to address this problem, consisting of (1) an unpaired generative adversarial network (GAN) for vendor-adaptation, and (2) a Unet for object segmentation. In the proposed Unet-GAN architecture, GAN learns from Unet at the feature level that is segmentation-specific. We used cardiac cine MRI as the example, with three major vendors (Philips, Siemens, and GE) as three domains, while the methodology can be extended to medical images segmentation in general. The proposed method showed significant improvement of the segmentation results across vendors. The proposed Unet-GAN provides an annotation-free solution to the cross-vendor medical image segmentation problem, potentially extending a trained deep learning model to multi-center and multi-vendor use in real clinical scenario.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge