Yaozong Gao
Cross-Site Severity Assessment of COVID-19 from CT Images via Domain Adaptation
Sep 08, 2021



Abstract:Early and accurate severity assessment of Coronavirus disease 2019 (COVID-19) based on computed tomography (CT) images offers a great help to the estimation of intensive care unit event and the clinical decision of treatment planning. To augment the labeled data and improve the generalization ability of the classification model, it is necessary to aggregate data from multiple sites. This task faces several challenges including class imbalance between mild and severe infections, domain distribution discrepancy between sites, and presence of heterogeneous features. In this paper, we propose a novel domain adaptation (DA) method with two components to address these problems. The first component is a stochastic class-balanced boosting sampling strategy that overcomes the imbalanced learning problem and improves the classification performance on poorly-predicted classes. The second component is a representation learning that guarantees three properties: 1) domain-transferability by prototype triplet loss, 2) discriminant by conditional maximum mean discrepancy loss, and 3) completeness by multi-view reconstruction loss. Particularly, we propose a domain translator and align the heterogeneous data to the estimated class prototypes (i.e., class centers) in a hyper-sphere manifold. Experiments on cross-site severity assessment of COVID-19 from CT images show that the proposed method can effectively tackle the imbalanced learning problem and outperform recent DA approaches.
Dual-Sampling Attention Network for Diagnosis of COVID-19 from Community Acquired Pneumonia
May 20, 2020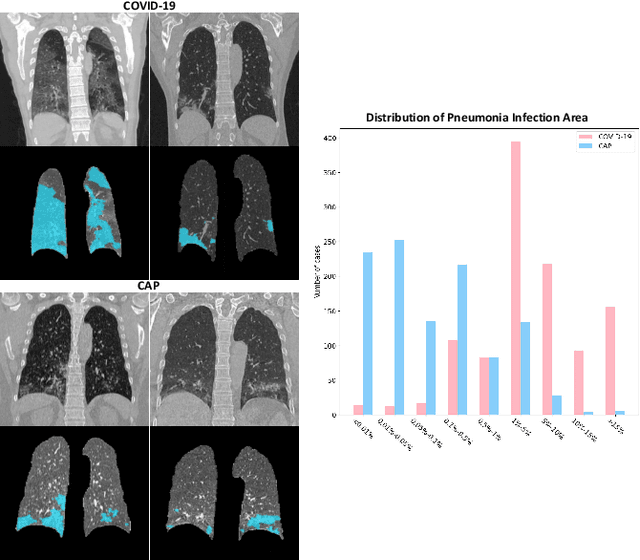
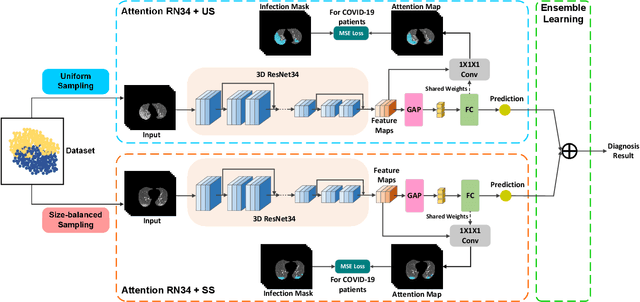
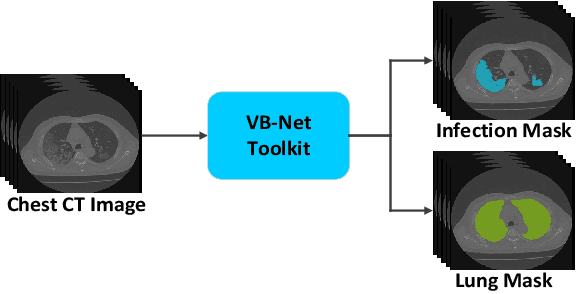
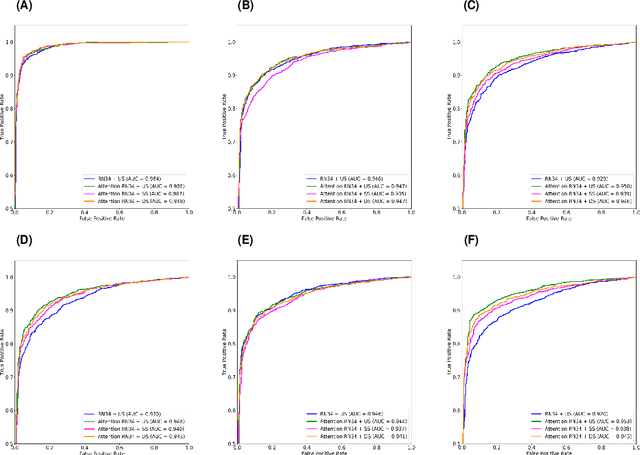
Abstract:The coronavirus disease (COVID-19) is rapidly spreading all over the world, and has infected more than 1,436,000 people in more than 200 countries and territories as of April 9, 2020. Detecting COVID-19 at early stage is essential to deliver proper healthcare to the patients and also to protect the uninfected population. To this end, we develop a dual-sampling attention network to automatically diagnose COVID- 19 from the community acquired pneumonia (CAP) in chest computed tomography (CT). In particular, we propose a novel online attention module with a 3D convolutional network (CNN) to focus on the infection regions in lungs when making decisions of diagnoses. Note that there exists imbalanced distribution of the sizes of the infection regions between COVID-19 and CAP, partially due to fast progress of COVID-19 after symptom onset. Therefore, we develop a dual-sampling strategy to mitigate the imbalanced learning. Our method is evaluated (to our best knowledge) upon the largest multi-center CT data for COVID-19 from 8 hospitals. In the training-validation stage, we collect 2186 CT scans from 1588 patients for a 5-fold cross-validation. In the testing stage, we employ another independent large-scale testing dataset including 2796 CT scans from 2057 patients. Results show that our algorithm can identify the COVID-19 images with the area under the receiver operating characteristic curve (AUC) value of 0.944, accuracy of 87.5%, sensitivity of 86.9%, specificity of 90.1%, and F1-score of 82.0%. With this performance, the proposed algorithm could potentially aid radiologists with COVID-19 diagnosis from CAP, especially in the early stage of the COVID-19 outbreak.
Joint Prediction and Time Estimation of COVID-19 Developing Severe Symptoms using Chest CT Scan
May 07, 2020
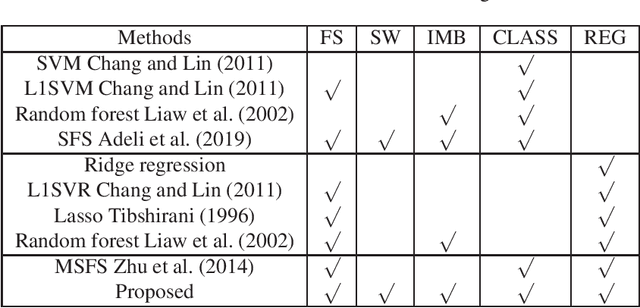
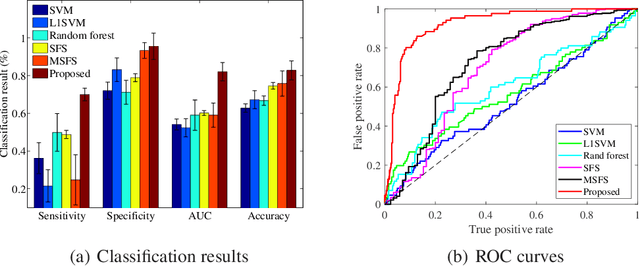
Abstract:With the rapidly worldwide spread of Coronavirus disease (COVID-19), it is of great importance to conduct early diagnosis of COVID-19 and predict the time that patients might convert to the severe stage, for designing effective treatment plan and reducing the clinicians' workloads. In this study, we propose a joint classification and regression method to determine whether the patient would develop severe symptoms in the later time, and if yes, predict the possible conversion time that the patient would spend to convert to the severe stage. To do this, the proposed method takes into account 1) the weight for each sample to reduce the outliers' influence and explore the problem of imbalance classification, and 2) the weight for each feature via a sparsity regularization term to remove the redundant features of high-dimensional data and learn the shared information across the classification task and the regression task. To our knowledge, this study is the first work to predict the disease progression and the conversion time, which could help clinicians to deal with the potential severe cases in time or even save the patients' lives. Experimental analysis was conducted on a real data set from two hospitals with 422 chest computed tomography (CT) scans, where 52 cases were converted to severe on average 5.64 days and 34 cases were severe at admission. Results show that our method achieves the best classification (e.g., 85.91% of accuracy) and regression (e.g., 0.462 of the correlation coefficient) performance, compared to all comparison methods. Moreover, our proposed method yields 76.97% of accuracy for predicting the severe cases, 0.524 of the correlation coefficient, and 0.55 days difference for the converted time.
Hypergraph Learning for Identification of COVID-19 with CT Imaging
May 07, 2020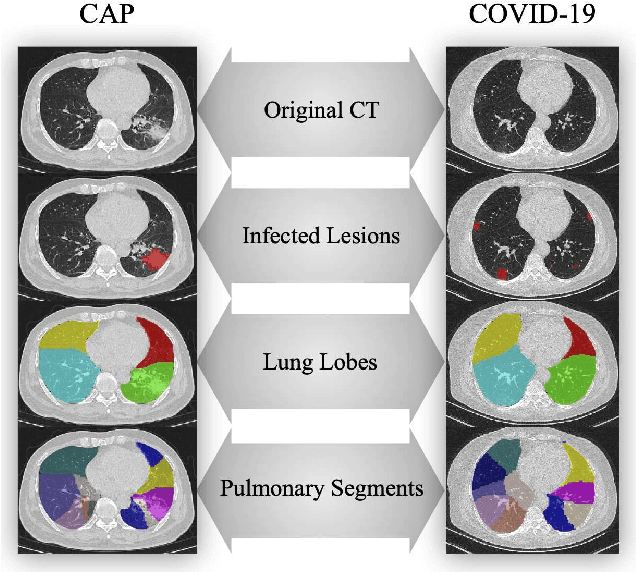
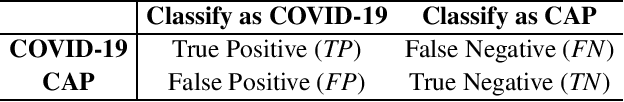
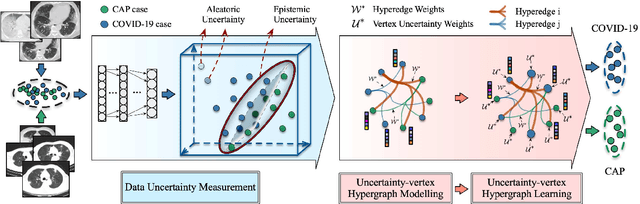
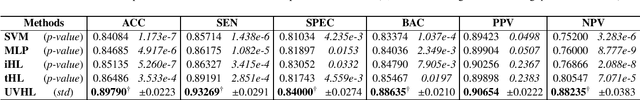
Abstract:The coronavirus disease, named COVID-19, has become the largest global public health crisis since it started in early 2020. CT imaging has been used as a complementary tool to assist early screening, especially for the rapid identification of COVID-19 cases from community acquired pneumonia (CAP) cases. The main challenge in early screening is how to model the confusing cases in the COVID-19 and CAP groups, with very similar clinical manifestations and imaging features. To tackle this challenge, we propose an Uncertainty Vertex-weighted Hypergraph Learning (UVHL) method to identify COVID-19 from CAP using CT images. In particular, multiple types of features (including regional features and radiomics features) are first extracted from CT image for each case. Then, the relationship among different cases is formulated by a hypergraph structure, with each case represented as a vertex in the hypergraph. The uncertainty of each vertex is further computed with an uncertainty score measurement and used as a weight in the hypergraph. Finally, a learning process of the vertex-weighted hypergraph is used to predict whether a new testing case belongs to COVID-19 or not. Experiments on a large multi-center pneumonia dataset, consisting of 2,148 COVID-19 cases and 1,182 CAP cases from five hospitals, are conducted to evaluate the performance of the proposed method. Results demonstrate the effectiveness and robustness of our proposed method on the identification of COVID-19 in comparison to state-of-the-art methods.
Adaptive Feature Selection Guided Deep Forest for COVID-19 Classification with Chest CT
May 07, 2020



Abstract:Chest computed tomography (CT) becomes an effective tool to assist the diagnosis of coronavirus disease-19 (COVID-19). Due to the outbreak of COVID-19 worldwide, using the computed-aided diagnosis technique for COVID-19 classification based on CT images could largely alleviate the burden of clinicians. In this paper, we propose an Adaptive Feature Selection guided Deep Forest (AFS-DF) for COVID-19 classification based on chest CT images. Specifically, we first extract location-specific features from CT images. Then, in order to capture the high-level representation of these features with the relatively small-scale data, we leverage a deep forest model to learn high-level representation of the features. Moreover, we propose a feature selection method based on the trained deep forest model to reduce the redundancy of features, where the feature selection could be adaptively incorporated with the COVID-19 classification model. We evaluated our proposed AFS-DF on COVID-19 dataset with 1495 patients of COVID-19 and 1027 patients of community acquired pneumonia (CAP). The accuracy (ACC), sensitivity (SEN), specificity (SPE) and AUC achieved by our method are 91.79%, 93.05%, 89.95% and 96.35%, respectively. Experimental results on the COVID-19 dataset suggest that the proposed AFS-DF achieves superior performance in COVID-19 vs. CAP classification, compared with 4 widely used machine learning methods.
Lung Infection Quantification of COVID-19 in CT Images with Deep Learning
Mar 30, 2020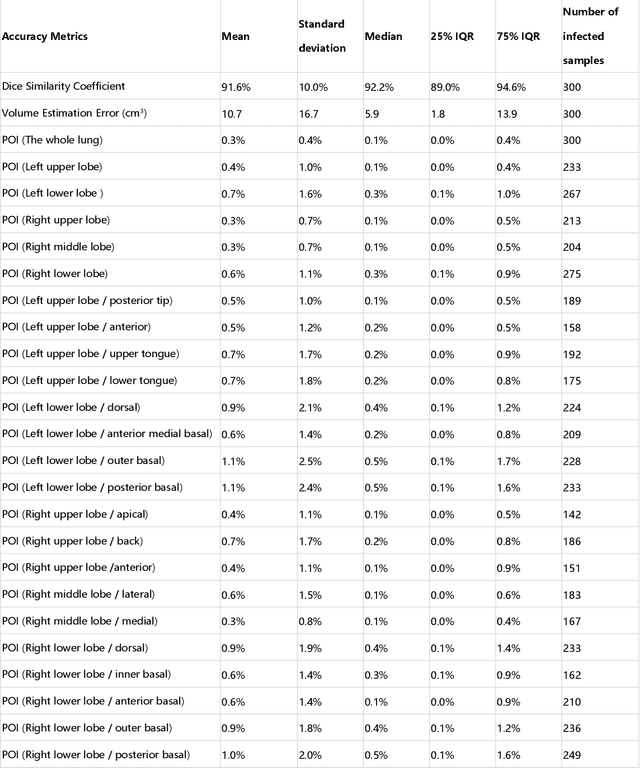
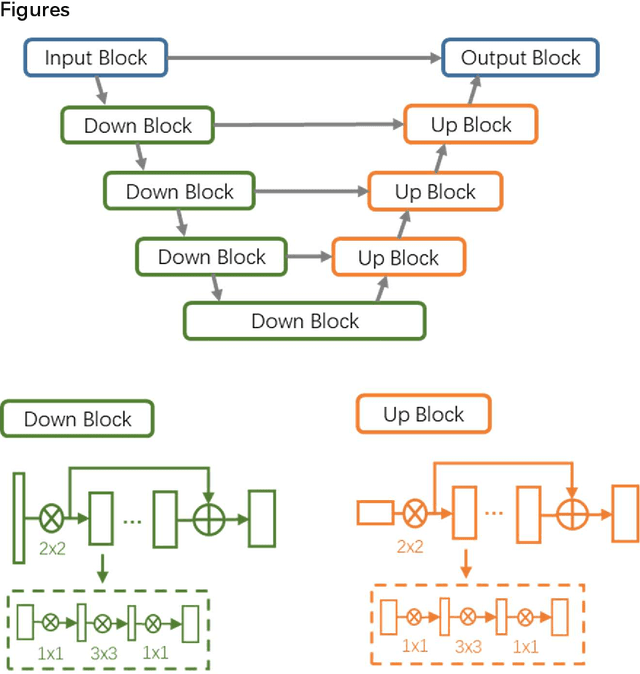
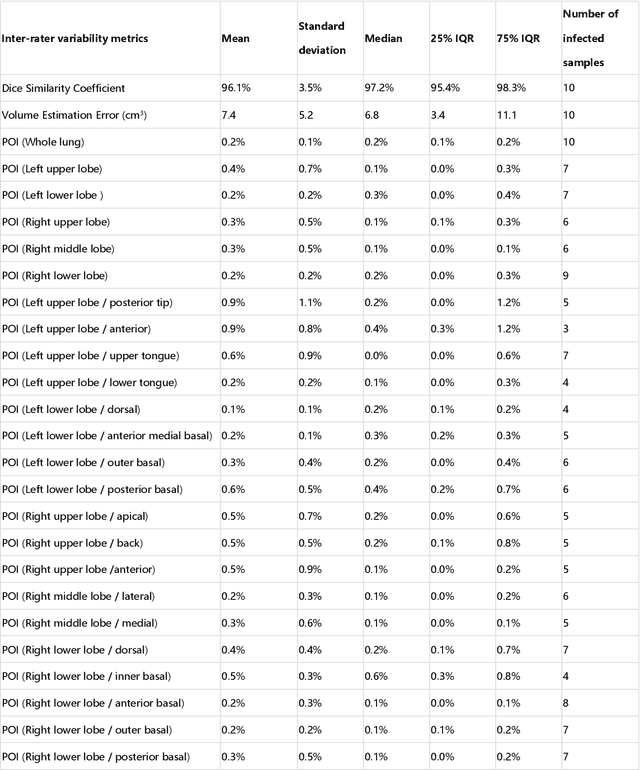
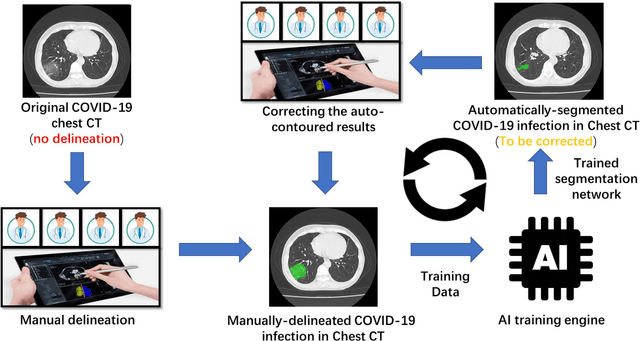
Abstract:CT imaging is crucial for diagnosis, assessment and staging COVID-19 infection. Follow-up scans every 3-5 days are often recommended for disease progression. It has been reported that bilateral and peripheral ground glass opacification (GGO) with or without consolidation are predominant CT findings in COVID-19 patients. However, due to lack of computerized quantification tools, only qualitative impression and rough description of infected areas are currently used in radiological reports. In this paper, a deep learning (DL)-based segmentation system is developed to automatically quantify infection regions of interest (ROIs) and their volumetric ratios w.r.t. the lung. The performance of the system was evaluated by comparing the automatically segmented infection regions with the manually-delineated ones on 300 chest CT scans of 300 COVID-19 patients. For fast manual delineation of training samples and possible manual intervention of automatic results, a human-in-the-loop (HITL) strategy has been adopted to assist radiologists for infection region segmentation, which dramatically reduced the total segmentation time to 4 minutes after 3 iterations of model updating. The average Dice simiarility coefficient showed 91.6% agreement between automatic and manual infaction segmentations, and the mean estimation error of percentage of infection (POI) was 0.3% for the whole lung. Finally, possible applications, including but not limited to analysis of follow-up CT scans and infection distributions in the lobes and segments correlated with clinical findings, were discussed.
Large-Scale Screening of COVID-19 from Community Acquired Pneumonia using Infection Size-Aware Classification
Mar 22, 2020



Abstract:The worldwide spread of coronavirus disease (COVID-19) has become a threatening risk for global public health. It is of great importance to rapidly and accurately screen patients with COVID-19 from community acquired pneumonia (CAP). In this study, a total of 1658 patients with COVID-19 and 1027 patients of CAP underwent thin-section CT. All images were preprocessed to obtain the segmentations of both infections and lung fields, which were used to extract location-specific features. An infection Size Aware Random Forest method (iSARF) was proposed, in which subjects were automated categorized into groups with different ranges of infected lesion sizes, followed by random forests in each group for classification. Experimental results show that the proposed method yielded sensitivity of 0.907, specificity of 0.833, and accuracy of 0.879 under five-fold cross-validation. Large performance margins against comparison methods were achieved especially for the cases with infection size in the medium range, from 0.01% to 10%. The further inclusion of Radiomics features show slightly improvement. It is anticipated that our proposed framework could assist clinical decision making.
The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: Results of the KiTS19 Challenge
Dec 02, 2019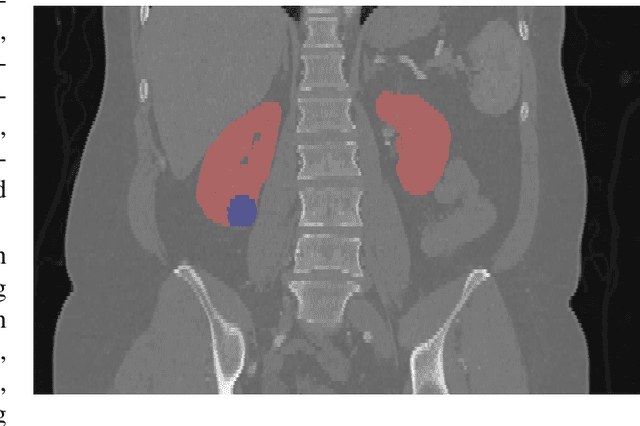
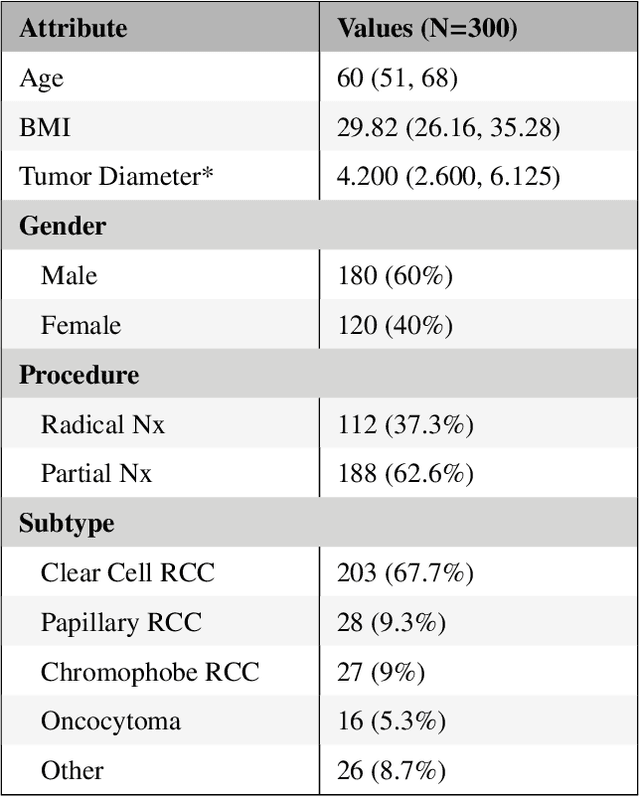
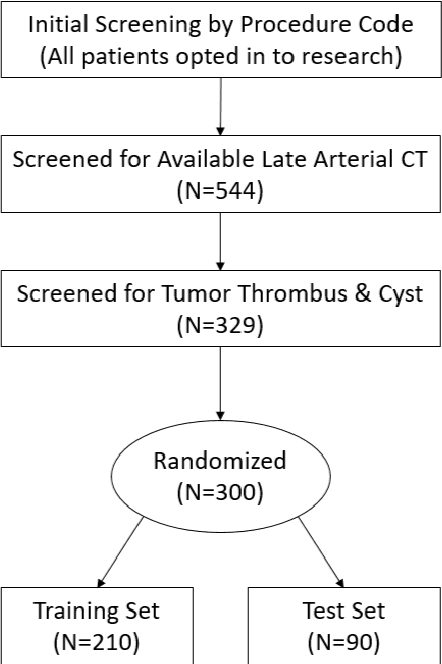
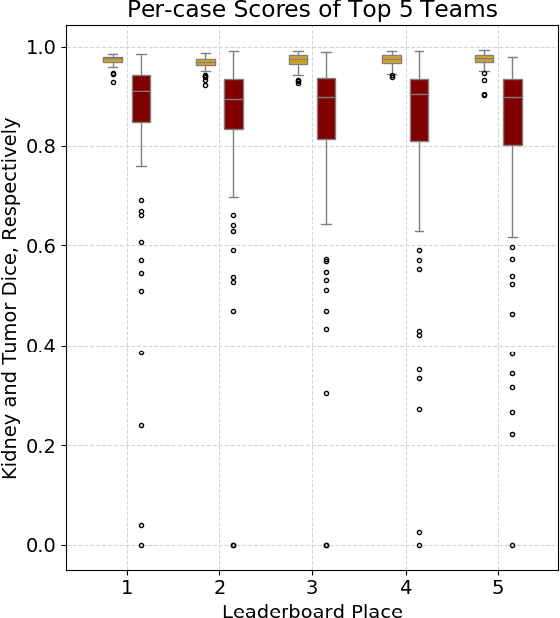
Abstract:There is a large body of literature linking anatomic and geometric characteristics of kidney tumors to perioperative and oncologic outcomes. Semantic segmentation of these tumors and their host kidneys is a promising tool for quantitatively characterizing these lesions, but its adoption is limited due to the manual effort required to produce high-quality 3D segmentations of these structures. Recently, methods based on deep learning have shown excellent results in automatic 3D segmentation, but they require large datasets for training, and there remains little consensus on which methods perform best. The 2019 Kidney and Kidney Tumor Segmentation challenge (KiTS19) was a competition held in conjunction with the 2019 International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) which sought to address these issues and stimulate progress on this automatic segmentation problem. A training set of 210 cross sectional CT images with kidney tumors was publicly released with corresponding semantic segmentation masks. 106 teams from five continents used this data to develop automated systems to predict the true segmentation masks on a test set of 90 CT images for which the corresponding ground truth segmentations were kept private. These predictions were scored and ranked according to their average So rensen-Dice coefficient between the kidney and tumor across all 90 cases. The winning team achieved a Dice of 0.974 for kidney and 0.851 for tumor, approaching the inter-annotator performance on kidney (0.983) but falling short on tumor (0.923). This challenge has now entered an "open leaderboard" phase where it serves as a challenging benchmark in 3D semantic segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge