Weiya Shi
X-Recon: Learning-based Patient-specific High-Resolution CT Reconstruction from Orthogonal X-Ray Images
Jul 22, 2024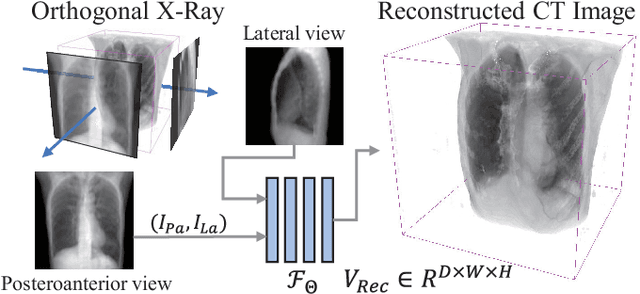
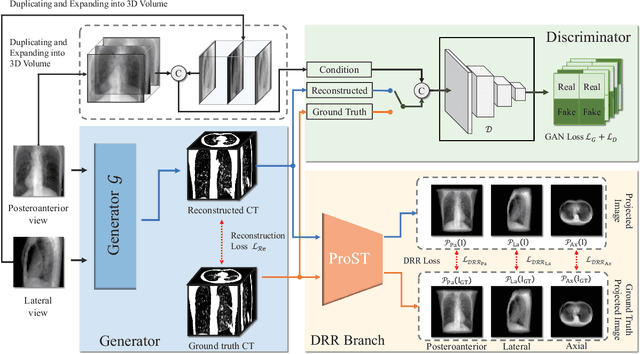
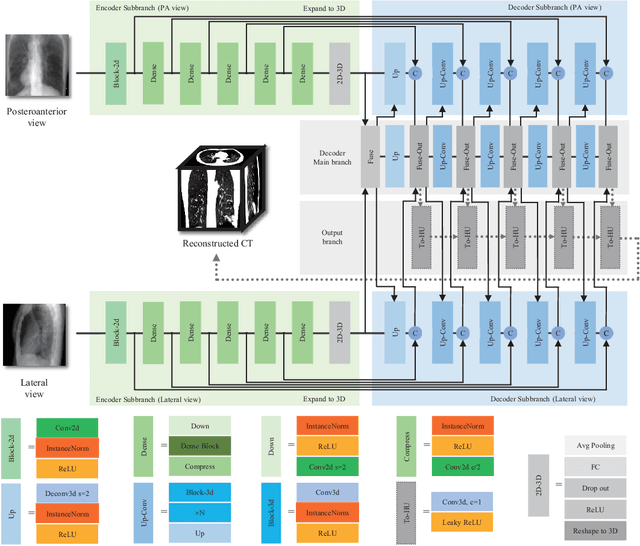
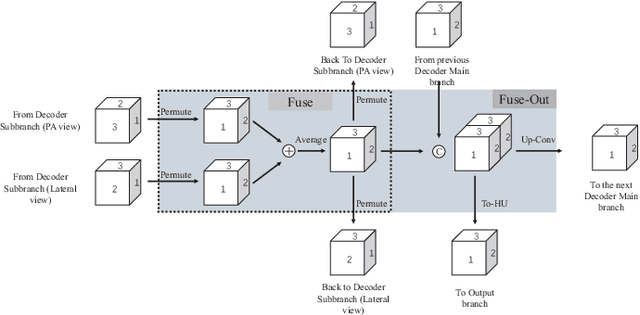
Abstract:Rapid and accurate diagnosis of pneumothorax, utilizing chest X-ray and computed tomography (CT), is crucial for assisted diagnosis. Chest X-ray is commonly used for initial localization of pneumothorax, while CT ensures accurate quantification. However, CT scans involve high radiation doses and can be costly. To achieve precise quantitative diagnosis while minimizing radiation exposure, we proposed X-Recon, a CT ultra-sparse reconstruction network based on ortho-lateral chest X-ray images. X-Recon integrates generative adversarial networks (GANs), including a generator with a multi-scale fusion rendering module and a discriminator enhanced by 3D coordinate convolutional layers, designed to facilitate CT reconstruction. To improve precision, a projective spatial transformer is utilized to incorporate multi-angle projection loss. Additionally, we proposed PTX-Seg, a zero-shot pneumothorax segmentation algorithm, combining image processing techniques with deep-learning models for the segmentation of air-accumulated regions and lung structures. Experiments on a large-scale dataset demonstrate its superiority over existing approaches. X-Recon achieved a significantly higher reconstruction resolution with a higher average spatial resolution and a lower average slice thickness. The reconstruction metrics achieved state-of-the-art performance in terms of several metrics including peak signal-to-noise ratio. The zero-shot segmentation algorithm, PTX-Seg, also demonstrated high segmentation precision for the air-accumulated region, the left lung, and the right lung. Moreover, the consistency analysis for the pneumothorax chest occupancy ratio between reconstructed CT and original CT obtained a high correlation coefficient. Code will be available at: https://github.com/wangyunpengbio/X-Recon
M3Lung-Sys: A Deep Learning System for Multi-Class Lung Pneumonia Screening from CT Imaging
Oct 07, 2020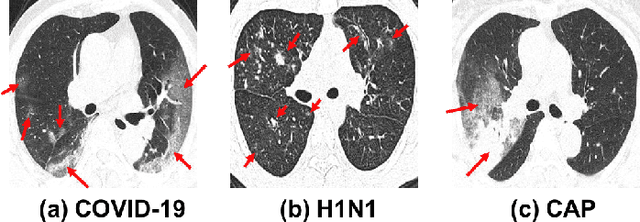
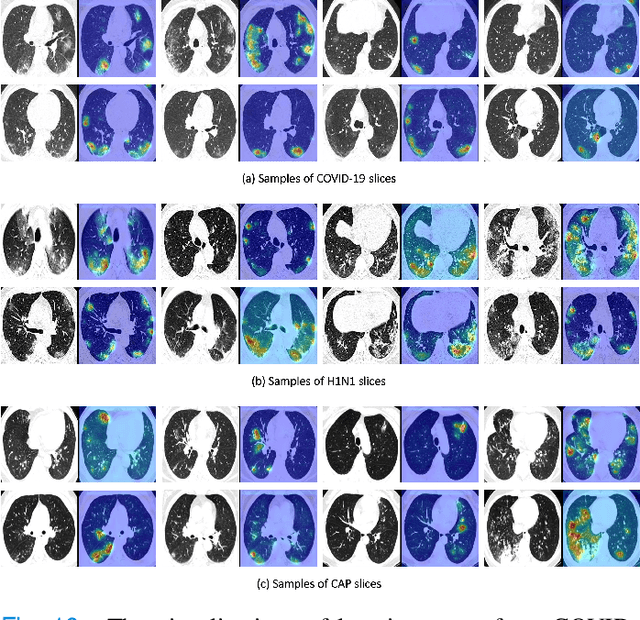
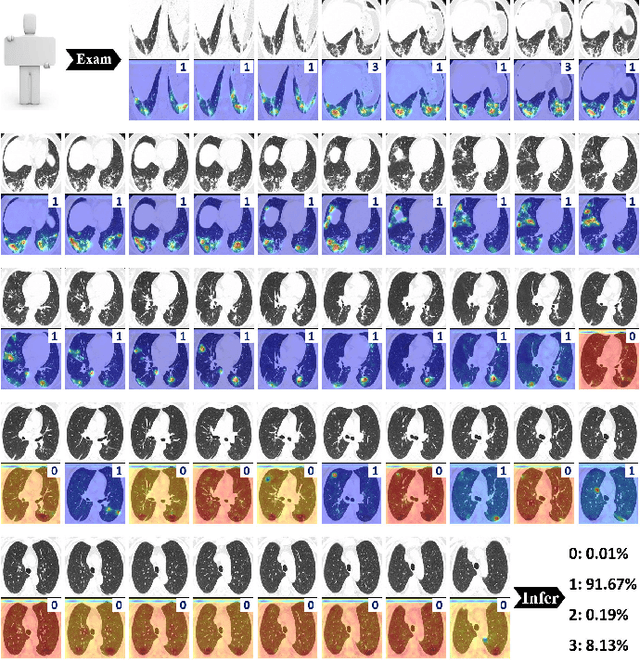
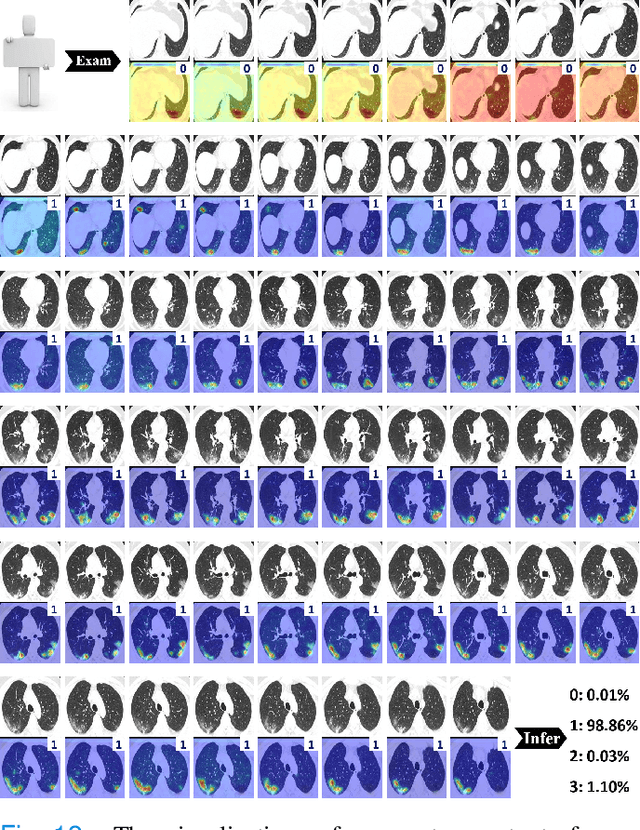
Abstract:To counter the outbreak of COVID-19, the accurate diagnosis of suspected cases plays a crucial role in timely quarantine, medical treatment, and preventing the spread of the pandemic. Considering the limited training cases and resources (e.g, time and budget), we propose a Multi-task Multi-slice Deep Learning System (M3Lung-Sys) for multi-class lung pneumonia screening from CT imaging, which only consists of two 2D CNN networks, i.e., slice- and patient-level classification networks. The former aims to seek the feature representations from abundant CT slices instead of limited CT volumes, and for the overall pneumonia screening, the latter one could recover the temporal information by feature refinement and aggregation between different slices. In addition to distinguish COVID-19 from Healthy, H1N1, and CAP cases, our M 3 Lung-Sys also be able to locate the areas of relevant lesions, without any pixel-level annotation. To further demonstrate the effectiveness of our model, we conduct extensive experiments on a chest CT imaging dataset with a total of 734 patients (251 healthy people, 245 COVID-19 patients, 105 H1N1 patients, and 133 CAP patients). The quantitative results with plenty of metrics indicate the superiority of our proposed model on both slice- and patient-level classification tasks. More importantly, the generated lesion location maps make our system interpretable and more valuable to clinicians.
Lung Infection Quantification of COVID-19 in CT Images with Deep Learning
Mar 30, 2020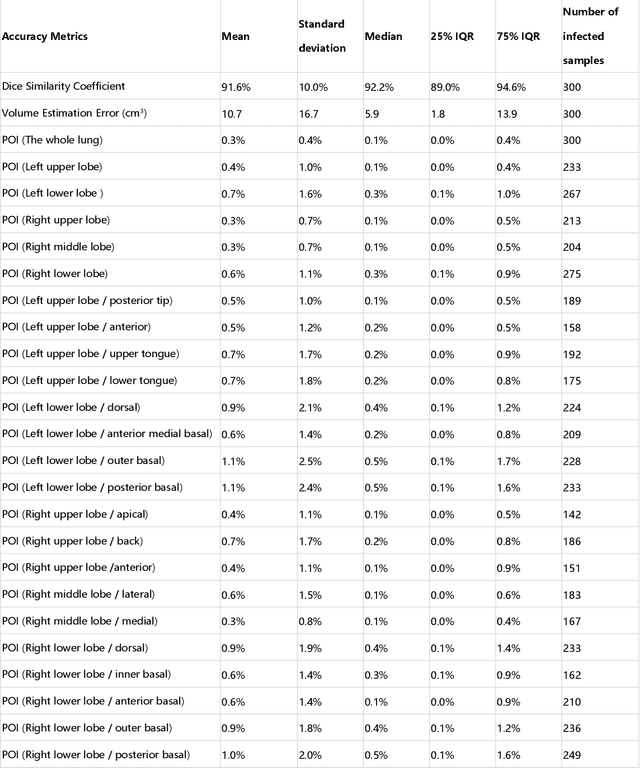
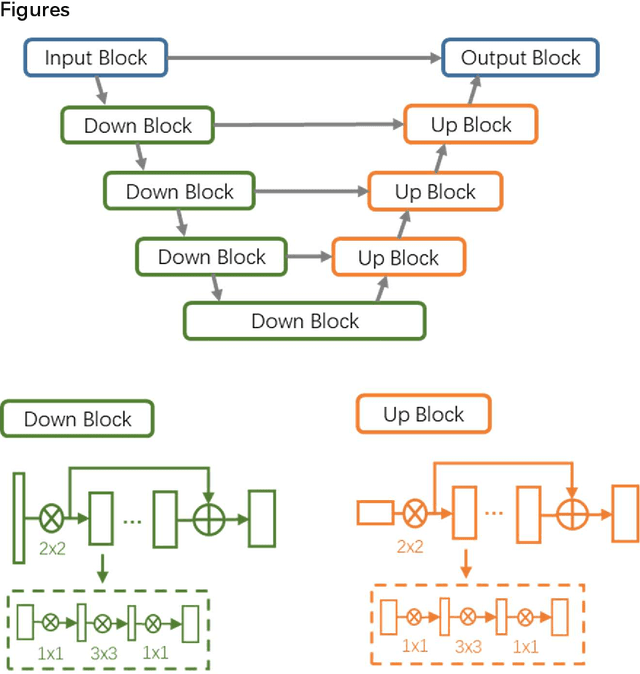
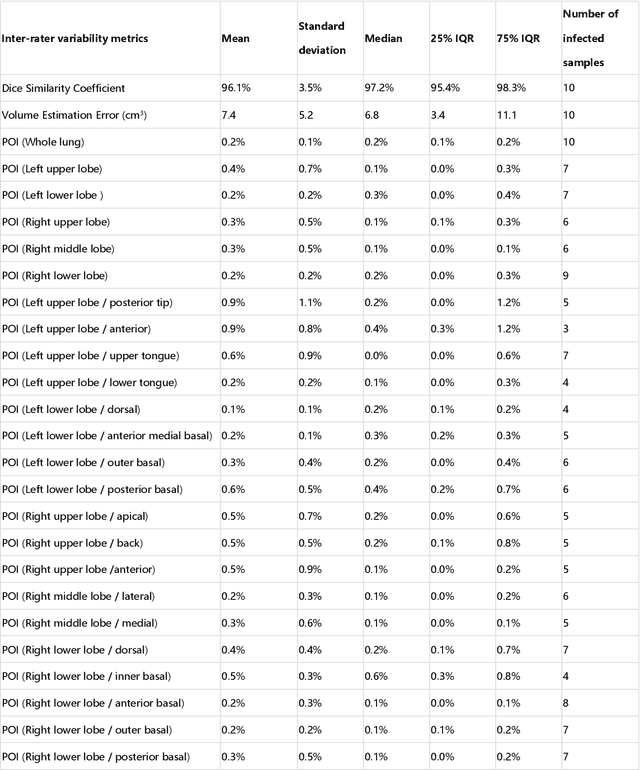
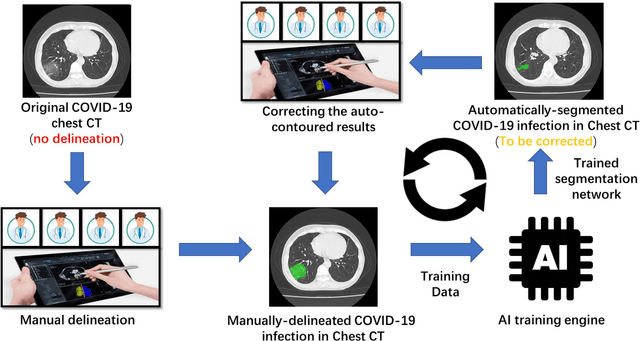
Abstract:CT imaging is crucial for diagnosis, assessment and staging COVID-19 infection. Follow-up scans every 3-5 days are often recommended for disease progression. It has been reported that bilateral and peripheral ground glass opacification (GGO) with or without consolidation are predominant CT findings in COVID-19 patients. However, due to lack of computerized quantification tools, only qualitative impression and rough description of infected areas are currently used in radiological reports. In this paper, a deep learning (DL)-based segmentation system is developed to automatically quantify infection regions of interest (ROIs) and their volumetric ratios w.r.t. the lung. The performance of the system was evaluated by comparing the automatically segmented infection regions with the manually-delineated ones on 300 chest CT scans of 300 COVID-19 patients. For fast manual delineation of training samples and possible manual intervention of automatic results, a human-in-the-loop (HITL) strategy has been adopted to assist radiologists for infection region segmentation, which dramatically reduced the total segmentation time to 4 minutes after 3 iterations of model updating. The average Dice simiarility coefficient showed 91.6% agreement between automatic and manual infaction segmentations, and the mean estimation error of percentage of infection (POI) was 0.3% for the whole lung. Finally, possible applications, including but not limited to analysis of follow-up CT scans and infection distributions in the lobes and segments correlated with clinical findings, were discussed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge