Fei Shan
X-Recon: Learning-based Patient-specific High-Resolution CT Reconstruction from Orthogonal X-Ray Images
Jul 22, 2024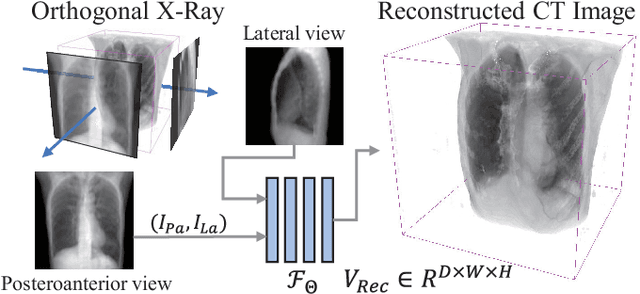
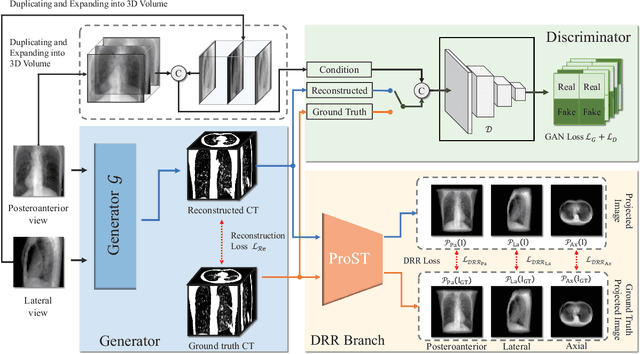
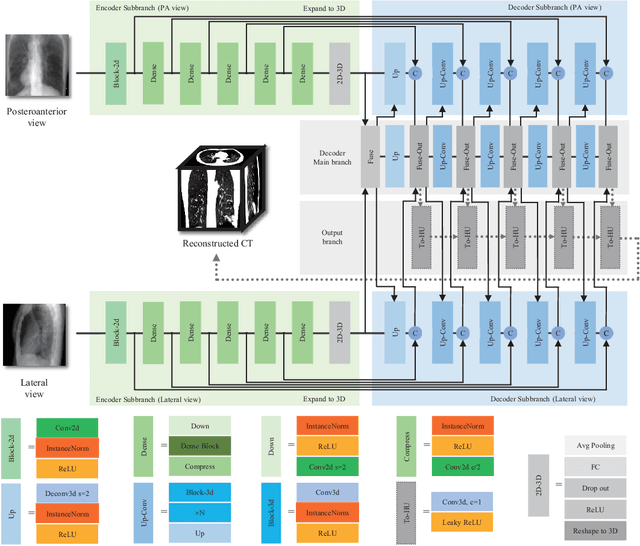
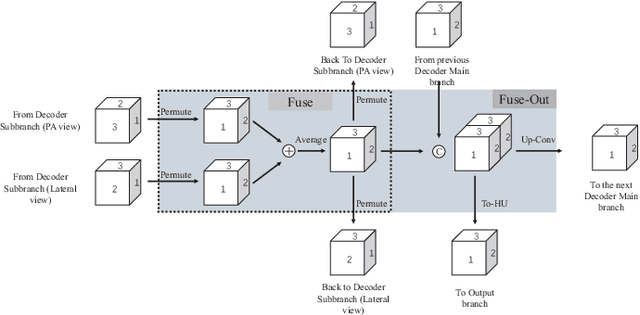
Abstract:Rapid and accurate diagnosis of pneumothorax, utilizing chest X-ray and computed tomography (CT), is crucial for assisted diagnosis. Chest X-ray is commonly used for initial localization of pneumothorax, while CT ensures accurate quantification. However, CT scans involve high radiation doses and can be costly. To achieve precise quantitative diagnosis while minimizing radiation exposure, we proposed X-Recon, a CT ultra-sparse reconstruction network based on ortho-lateral chest X-ray images. X-Recon integrates generative adversarial networks (GANs), including a generator with a multi-scale fusion rendering module and a discriminator enhanced by 3D coordinate convolutional layers, designed to facilitate CT reconstruction. To improve precision, a projective spatial transformer is utilized to incorporate multi-angle projection loss. Additionally, we proposed PTX-Seg, a zero-shot pneumothorax segmentation algorithm, combining image processing techniques with deep-learning models for the segmentation of air-accumulated regions and lung structures. Experiments on a large-scale dataset demonstrate its superiority over existing approaches. X-Recon achieved a significantly higher reconstruction resolution with a higher average spatial resolution and a lower average slice thickness. The reconstruction metrics achieved state-of-the-art performance in terms of several metrics including peak signal-to-noise ratio. The zero-shot segmentation algorithm, PTX-Seg, also demonstrated high segmentation precision for the air-accumulated region, the left lung, and the right lung. Moreover, the consistency analysis for the pneumothorax chest occupancy ratio between reconstructed CT and original CT obtained a high correlation coefficient. Code will be available at: https://github.com/wangyunpengbio/X-Recon
Towards Unified Surgical Skill Assessment
Jun 02, 2021



Abstract:Surgical skills have a great influence on surgical safety and patients' well-being. Traditional assessment of surgical skills involves strenuous manual efforts, which lacks efficiency and repeatability. Therefore, we attempt to automatically predict how well the surgery is performed using the surgical video. In this paper, a unified multi-path framework for automatic surgical skill assessment is proposed, which takes care of multiple composing aspects of surgical skills, including surgical tool usage, intraoperative event pattern, and other skill proxies. The dependency relationships among these different aspects are specially modeled by a path dependency module in the framework. We conduct extensive experiments on the JIGSAWS dataset of simulated surgical tasks, and a new clinical dataset of real laparoscopic surgeries. The proposed framework achieves promising results on both datasets, with the state-of-the-art on the simulated dataset advanced from 0.71 Spearman's correlation to 0.80. It is also shown that combining multiple skill aspects yields better performance than relying on a single aspect.
M3Lung-Sys: A Deep Learning System for Multi-Class Lung Pneumonia Screening from CT Imaging
Oct 07, 2020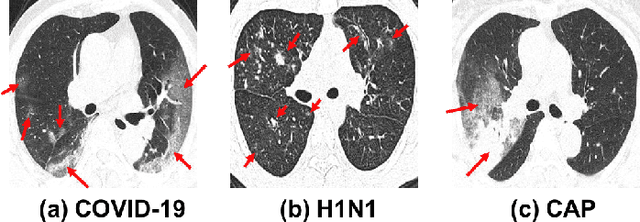
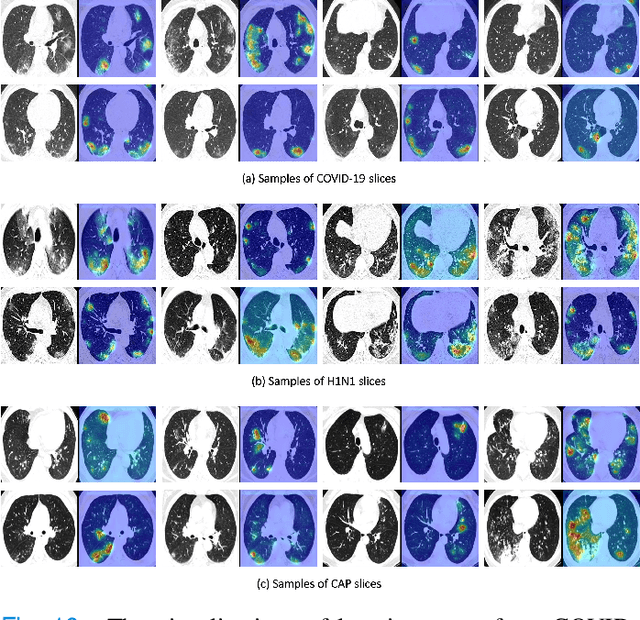
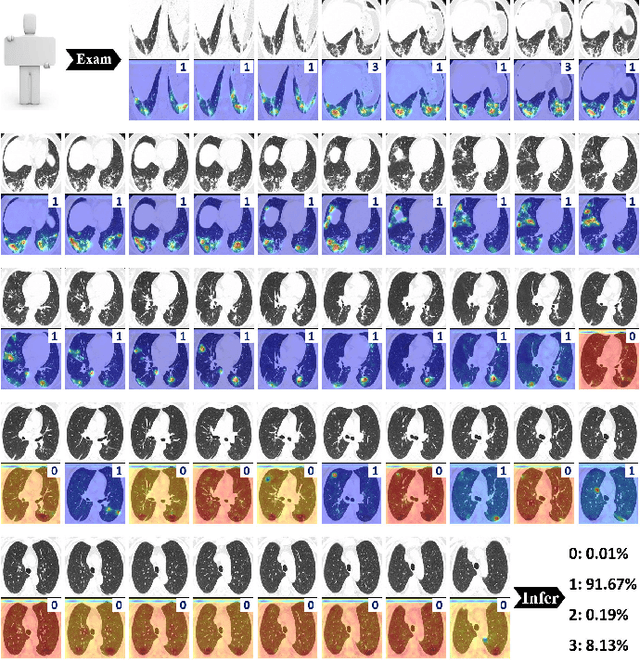
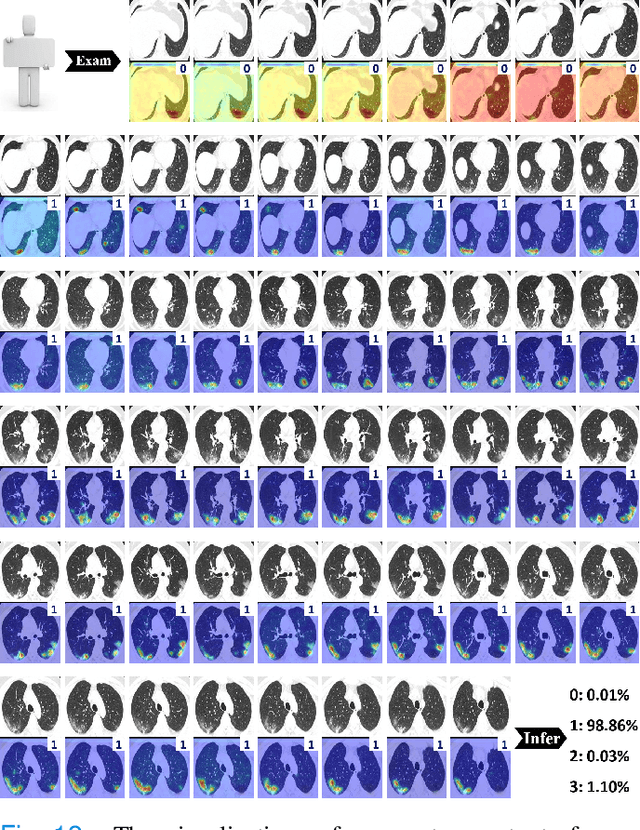
Abstract:To counter the outbreak of COVID-19, the accurate diagnosis of suspected cases plays a crucial role in timely quarantine, medical treatment, and preventing the spread of the pandemic. Considering the limited training cases and resources (e.g, time and budget), we propose a Multi-task Multi-slice Deep Learning System (M3Lung-Sys) for multi-class lung pneumonia screening from CT imaging, which only consists of two 2D CNN networks, i.e., slice- and patient-level classification networks. The former aims to seek the feature representations from abundant CT slices instead of limited CT volumes, and for the overall pneumonia screening, the latter one could recover the temporal information by feature refinement and aggregation between different slices. In addition to distinguish COVID-19 from Healthy, H1N1, and CAP cases, our M 3 Lung-Sys also be able to locate the areas of relevant lesions, without any pixel-level annotation. To further demonstrate the effectiveness of our model, we conduct extensive experiments on a chest CT imaging dataset with a total of 734 patients (251 healthy people, 245 COVID-19 patients, 105 H1N1 patients, and 133 CAP patients). The quantitative results with plenty of metrics indicate the superiority of our proposed model on both slice- and patient-level classification tasks. More importantly, the generated lesion location maps make our system interpretable and more valuable to clinicians.
Surgical Skill Assessment on In-Vivo Clinical Data via the Clearness of Operating Field
Aug 27, 2020



Abstract:Surgical skill assessment is important for surgery training and quality control. Prior works on this task largely focus on basic surgical tasks such as suturing and knot tying performed in simulation settings. In contrast, surgical skill assessment is studied in this paper on a real clinical dataset, which consists of fifty-seven in-vivo laparoscopic surgeries and corresponding skill scores annotated by six surgeons. From analyses on this dataset, the clearness of operating field (COF) is identified as a good proxy for overall surgical skills, given its strong correlation with overall skills and high inter-annotator consistency. Then an objective and automated framework based on neural network is proposed to predict surgical skills through the proxy of COF. The neural network is jointly trained with a supervised regression loss and an unsupervised rank loss. In experiments, the proposed method achieves 0.55 Spearman's correlation with the ground truth of overall technical skill, which is even comparable with the human performance of junior surgeons.
Unsupervised Surgical Instrument Segmentation via Anchor Generation and Semantic Diffusion
Aug 27, 2020



Abstract:Surgical instrument segmentation is a key component in developing context-aware operating rooms. Existing works on this task heavily rely on the supervision of a large amount of labeled data, which involve laborious and expensive human efforts. In contrast, a more affordable unsupervised approach is developed in this paper. To train our model, we first generate anchors as pseudo labels for instruments and background tissues respectively by fusing coarse handcrafted cues. Then a semantic diffusion loss is proposed to resolve the ambiguity in the generated anchors via the feature correlation between adjacent video frames. In the experiments on the binary instrument segmentation task of the 2017 MICCAI EndoVis Robotic Instrument Segmentation Challenge dataset, the proposed method achieves 0.71 IoU and 0.81 Dice score without using a single manual annotation, which is promising to show the potential of unsupervised learning for surgical tool segmentation.
Dual-Sampling Attention Network for Diagnosis of COVID-19 from Community Acquired Pneumonia
May 20, 2020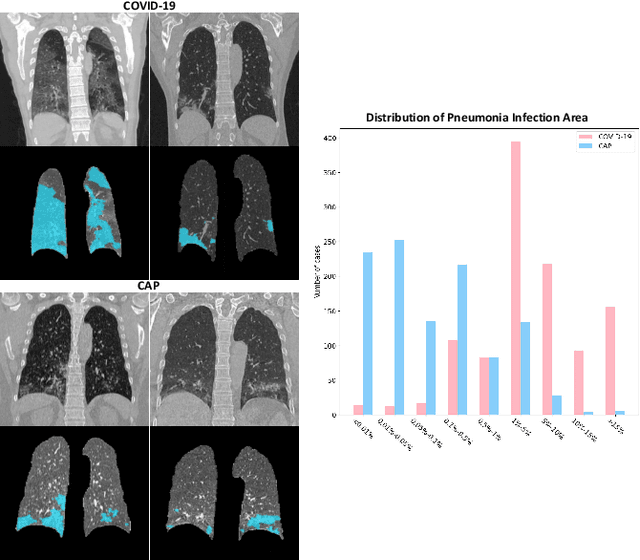
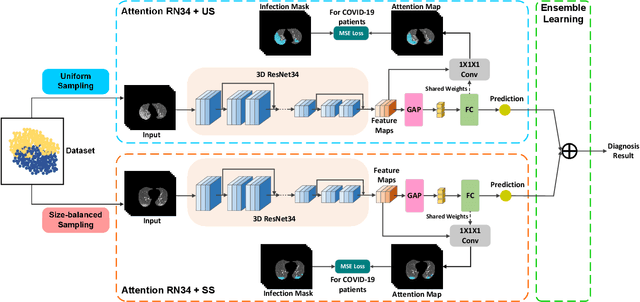
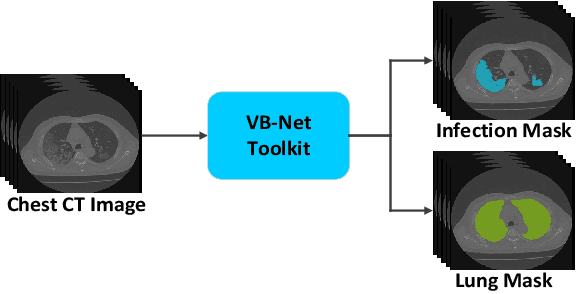
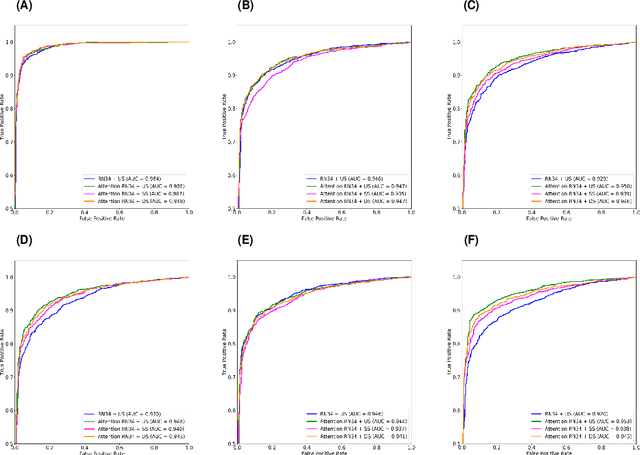
Abstract:The coronavirus disease (COVID-19) is rapidly spreading all over the world, and has infected more than 1,436,000 people in more than 200 countries and territories as of April 9, 2020. Detecting COVID-19 at early stage is essential to deliver proper healthcare to the patients and also to protect the uninfected population. To this end, we develop a dual-sampling attention network to automatically diagnose COVID- 19 from the community acquired pneumonia (CAP) in chest computed tomography (CT). In particular, we propose a novel online attention module with a 3D convolutional network (CNN) to focus on the infection regions in lungs when making decisions of diagnoses. Note that there exists imbalanced distribution of the sizes of the infection regions between COVID-19 and CAP, partially due to fast progress of COVID-19 after symptom onset. Therefore, we develop a dual-sampling strategy to mitigate the imbalanced learning. Our method is evaluated (to our best knowledge) upon the largest multi-center CT data for COVID-19 from 8 hospitals. In the training-validation stage, we collect 2186 CT scans from 1588 patients for a 5-fold cross-validation. In the testing stage, we employ another independent large-scale testing dataset including 2796 CT scans from 2057 patients. Results show that our algorithm can identify the COVID-19 images with the area under the receiver operating characteristic curve (AUC) value of 0.944, accuracy of 87.5%, sensitivity of 86.9%, specificity of 90.1%, and F1-score of 82.0%. With this performance, the proposed algorithm could potentially aid radiologists with COVID-19 diagnosis from CAP, especially in the early stage of the COVID-19 outbreak.
Joint Prediction and Time Estimation of COVID-19 Developing Severe Symptoms using Chest CT Scan
May 07, 2020
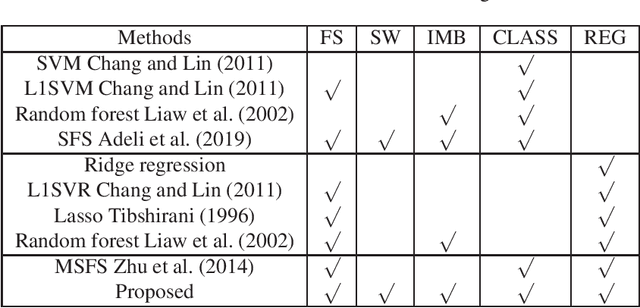
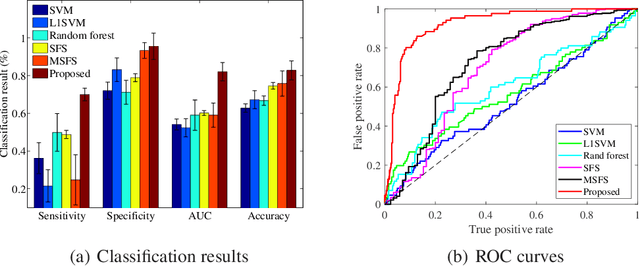
Abstract:With the rapidly worldwide spread of Coronavirus disease (COVID-19), it is of great importance to conduct early diagnosis of COVID-19 and predict the time that patients might convert to the severe stage, for designing effective treatment plan and reducing the clinicians' workloads. In this study, we propose a joint classification and regression method to determine whether the patient would develop severe symptoms in the later time, and if yes, predict the possible conversion time that the patient would spend to convert to the severe stage. To do this, the proposed method takes into account 1) the weight for each sample to reduce the outliers' influence and explore the problem of imbalance classification, and 2) the weight for each feature via a sparsity regularization term to remove the redundant features of high-dimensional data and learn the shared information across the classification task and the regression task. To our knowledge, this study is the first work to predict the disease progression and the conversion time, which could help clinicians to deal with the potential severe cases in time or even save the patients' lives. Experimental analysis was conducted on a real data set from two hospitals with 422 chest computed tomography (CT) scans, where 52 cases were converted to severe on average 5.64 days and 34 cases were severe at admission. Results show that our method achieves the best classification (e.g., 85.91% of accuracy) and regression (e.g., 0.462 of the correlation coefficient) performance, compared to all comparison methods. Moreover, our proposed method yields 76.97% of accuracy for predicting the severe cases, 0.524 of the correlation coefficient, and 0.55 days difference for the converted time.
Hypergraph Learning for Identification of COVID-19 with CT Imaging
May 07, 2020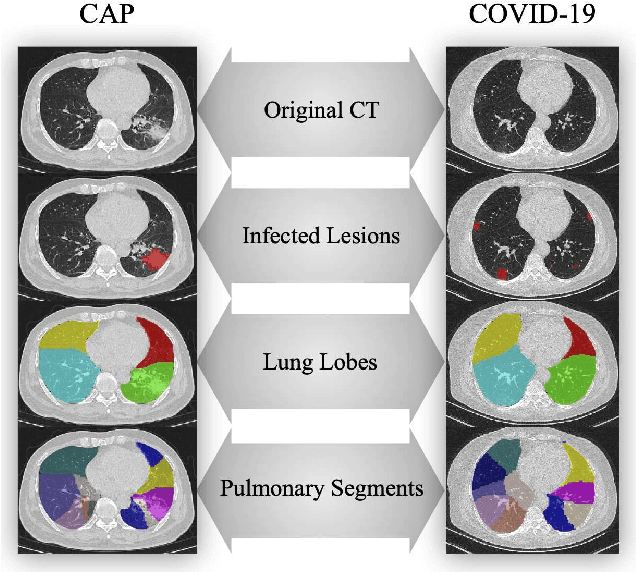
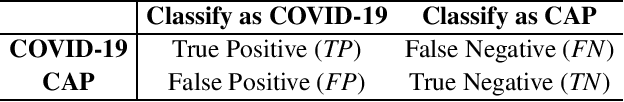
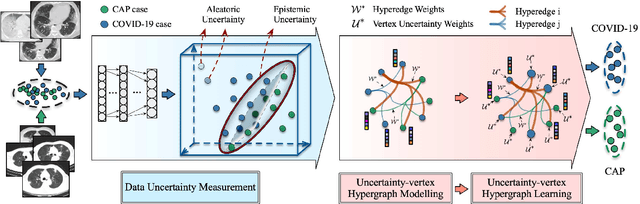
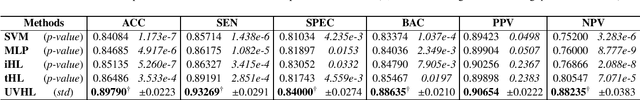
Abstract:The coronavirus disease, named COVID-19, has become the largest global public health crisis since it started in early 2020. CT imaging has been used as a complementary tool to assist early screening, especially for the rapid identification of COVID-19 cases from community acquired pneumonia (CAP) cases. The main challenge in early screening is how to model the confusing cases in the COVID-19 and CAP groups, with very similar clinical manifestations and imaging features. To tackle this challenge, we propose an Uncertainty Vertex-weighted Hypergraph Learning (UVHL) method to identify COVID-19 from CAP using CT images. In particular, multiple types of features (including regional features and radiomics features) are first extracted from CT image for each case. Then, the relationship among different cases is formulated by a hypergraph structure, with each case represented as a vertex in the hypergraph. The uncertainty of each vertex is further computed with an uncertainty score measurement and used as a weight in the hypergraph. Finally, a learning process of the vertex-weighted hypergraph is used to predict whether a new testing case belongs to COVID-19 or not. Experiments on a large multi-center pneumonia dataset, consisting of 2,148 COVID-19 cases and 1,182 CAP cases from five hospitals, are conducted to evaluate the performance of the proposed method. Results demonstrate the effectiveness and robustness of our proposed method on the identification of COVID-19 in comparison to state-of-the-art methods.
Adaptive Feature Selection Guided Deep Forest for COVID-19 Classification with Chest CT
May 07, 2020



Abstract:Chest computed tomography (CT) becomes an effective tool to assist the diagnosis of coronavirus disease-19 (COVID-19). Due to the outbreak of COVID-19 worldwide, using the computed-aided diagnosis technique for COVID-19 classification based on CT images could largely alleviate the burden of clinicians. In this paper, we propose an Adaptive Feature Selection guided Deep Forest (AFS-DF) for COVID-19 classification based on chest CT images. Specifically, we first extract location-specific features from CT images. Then, in order to capture the high-level representation of these features with the relatively small-scale data, we leverage a deep forest model to learn high-level representation of the features. Moreover, we propose a feature selection method based on the trained deep forest model to reduce the redundancy of features, where the feature selection could be adaptively incorporated with the COVID-19 classification model. We evaluated our proposed AFS-DF on COVID-19 dataset with 1495 patients of COVID-19 and 1027 patients of community acquired pneumonia (CAP). The accuracy (ACC), sensitivity (SEN), specificity (SPE) and AUC achieved by our method are 91.79%, 93.05%, 89.95% and 96.35%, respectively. Experimental results on the COVID-19 dataset suggest that the proposed AFS-DF achieves superior performance in COVID-19 vs. CAP classification, compared with 4 widely used machine learning methods.
Lung Infection Quantification of COVID-19 in CT Images with Deep Learning
Mar 30, 2020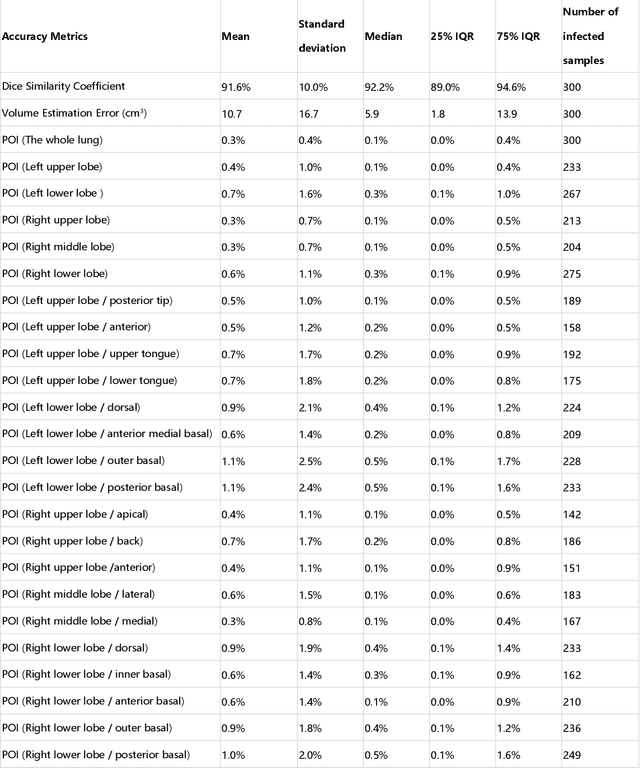
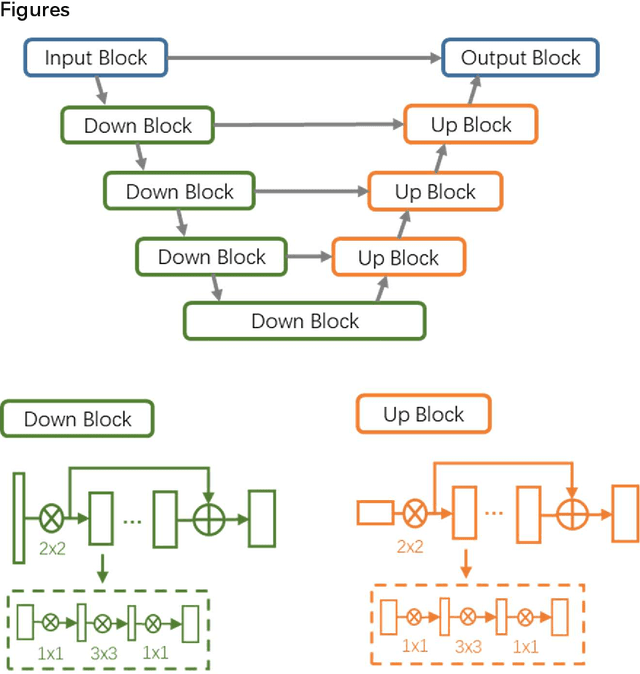
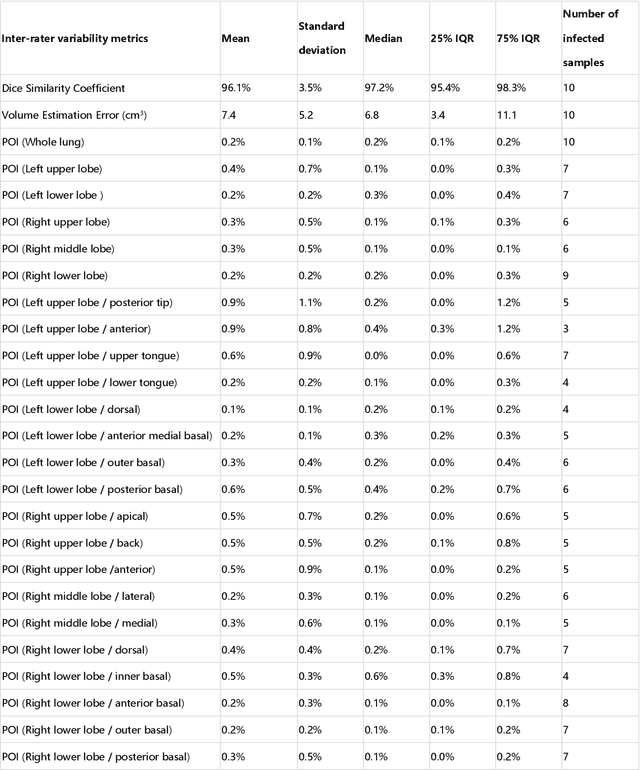
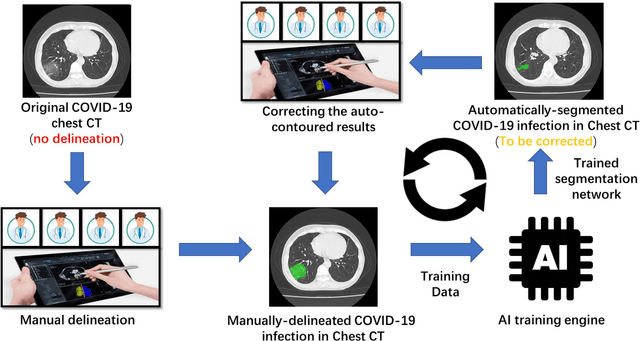
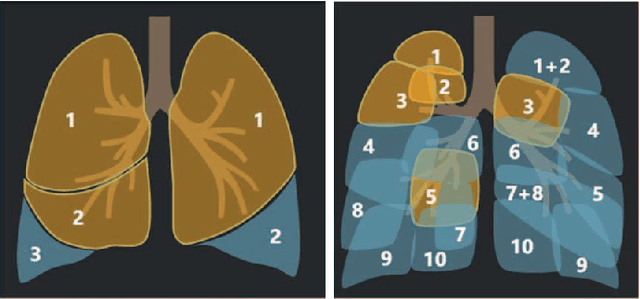
Abstract:CT imaging is crucial for diagnosis, assessment and staging COVID-19 infection. Follow-up scans every 3-5 days are often recommended for disease progression. It has been reported that bilateral and peripheral ground glass opacification (GGO) with or without consolidation are predominant CT findings in COVID-19 patients. However, due to lack of computerized quantification tools, only qualitative impression and rough description of infected areas are currently used in radiological reports. In this paper, a deep learning (DL)-based segmentation system is developed to automatically quantify infection regions of interest (ROIs) and their volumetric ratios w.r.t. the lung. The performance of the system was evaluated by comparing the automatically segmented infection regions with the manually-delineated ones on 300 chest CT scans of 300 COVID-19 patients. For fast manual delineation of training samples and possible manual intervention of automatic results, a human-in-the-loop (HITL) strategy has been adopted to assist radiologists for infection region segmentation, which dramatically reduced the total segmentation time to 4 minutes after 3 iterations of model updating. The average Dice simiarility coefficient showed 91.6% agreement between automatic and manual infaction segmentations, and the mean estimation error of percentage of infection (POI) was 0.3% for the whole lung. Finally, possible applications, including but not limited to analysis of follow-up CT scans and infection distributions in the lobes and segments correlated with clinical findings, were discussed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge