Miaofei Han
Hypergraph Learning for Identification of COVID-19 with CT Imaging
May 07, 2020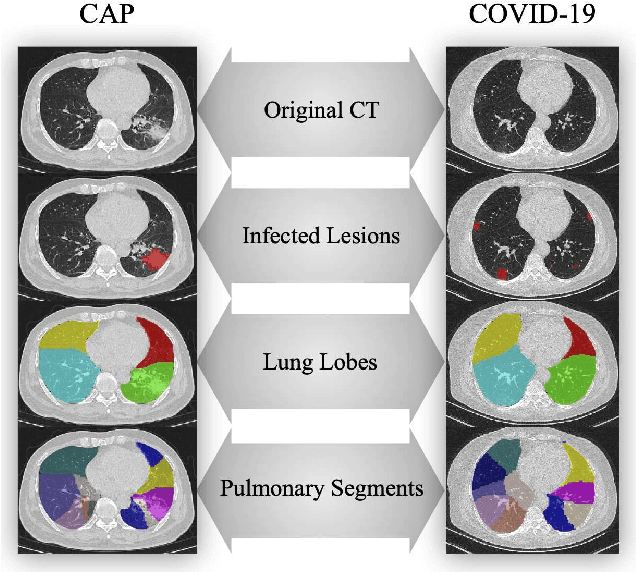
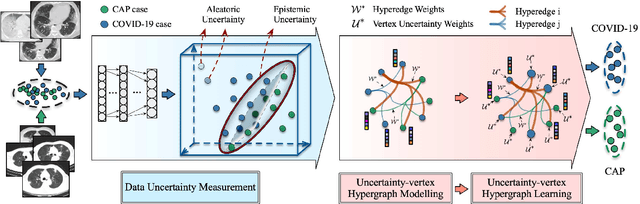
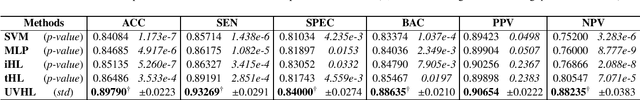
Abstract:The coronavirus disease, named COVID-19, has become the largest global public health crisis since it started in early 2020. CT imaging has been used as a complementary tool to assist early screening, especially for the rapid identification of COVID-19 cases from community acquired pneumonia (CAP) cases. The main challenge in early screening is how to model the confusing cases in the COVID-19 and CAP groups, with very similar clinical manifestations and imaging features. To tackle this challenge, we propose an Uncertainty Vertex-weighted Hypergraph Learning (UVHL) method to identify COVID-19 from CAP using CT images. In particular, multiple types of features (including regional features and radiomics features) are first extracted from CT image for each case. Then, the relationship among different cases is formulated by a hypergraph structure, with each case represented as a vertex in the hypergraph. The uncertainty of each vertex is further computed with an uncertainty score measurement and used as a weight in the hypergraph. Finally, a learning process of the vertex-weighted hypergraph is used to predict whether a new testing case belongs to COVID-19 or not. Experiments on a large multi-center pneumonia dataset, consisting of 2,148 COVID-19 cases and 1,182 CAP cases from five hospitals, are conducted to evaluate the performance of the proposed method. Results demonstrate the effectiveness and robustness of our proposed method on the identification of COVID-19 in comparison to state-of-the-art methods.
Lung Infection Quantification of COVID-19 in CT Images with Deep Learning
Mar 30, 2020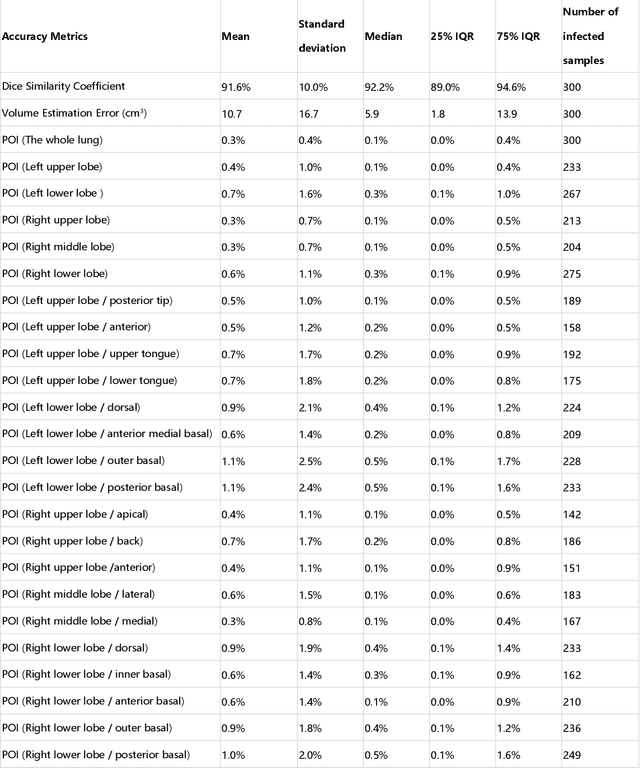
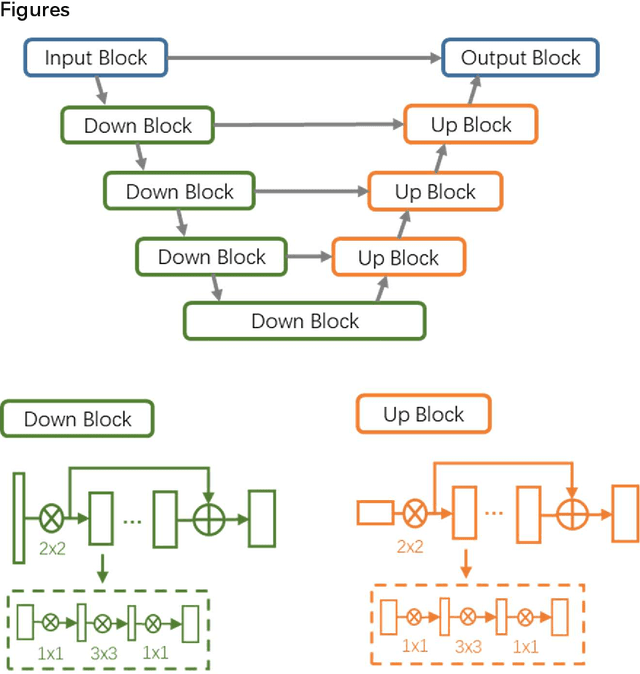
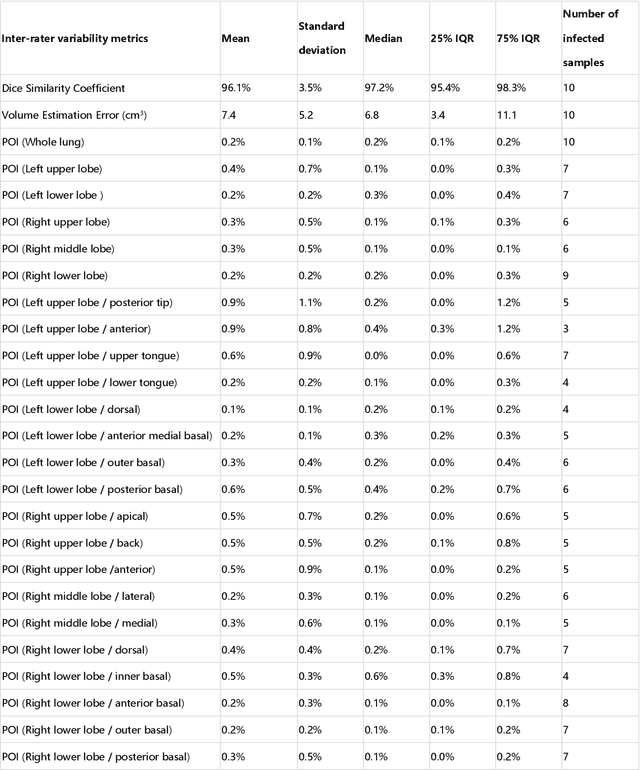
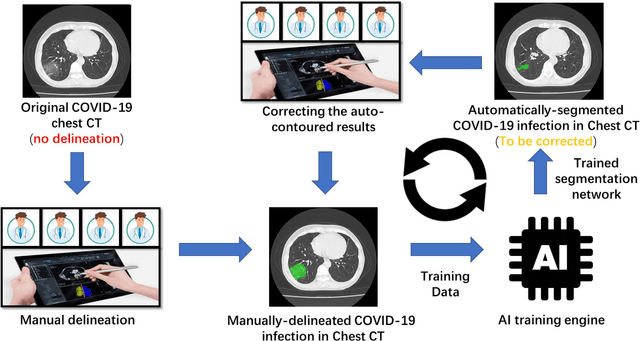
Abstract:CT imaging is crucial for diagnosis, assessment and staging COVID-19 infection. Follow-up scans every 3-5 days are often recommended for disease progression. It has been reported that bilateral and peripheral ground glass opacification (GGO) with or without consolidation are predominant CT findings in COVID-19 patients. However, due to lack of computerized quantification tools, only qualitative impression and rough description of infected areas are currently used in radiological reports. In this paper, a deep learning (DL)-based segmentation system is developed to automatically quantify infection regions of interest (ROIs) and their volumetric ratios w.r.t. the lung. The performance of the system was evaluated by comparing the automatically segmented infection regions with the manually-delineated ones on 300 chest CT scans of 300 COVID-19 patients. For fast manual delineation of training samples and possible manual intervention of automatic results, a human-in-the-loop (HITL) strategy has been adopted to assist radiologists for infection region segmentation, which dramatically reduced the total segmentation time to 4 minutes after 3 iterations of model updating. The average Dice simiarility coefficient showed 91.6% agreement between automatic and manual infaction segmentations, and the mean estimation error of percentage of infection (POI) was 0.3% for the whole lung. Finally, possible applications, including but not limited to analysis of follow-up CT scans and infection distributions in the lobes and segments correlated with clinical findings, were discussed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge