Kang Wang
Early and Prediagnostic Detection of Pancreatic Cancer from Computed Tomography
Jan 29, 2026Abstract:Pancreatic ductal adenocarcinoma (PDAC), one of the deadliest solid malignancies, is often detected at a late and inoperable stage. Retrospective reviews of prediagnostic CT scans, when conducted by expert radiologists aware that the patient later developed PDAC, frequently reveal lesions that were previously overlooked. To help detecting these lesions earlier, we developed an automated system named ePAI (early Pancreatic cancer detection with Artificial Intelligence). It was trained on data from 1,598 patients from a single medical center. In the internal test involving 1,009 patients, ePAI achieved an area under the receiver operating characteristic curve (AUC) of 0.939-0.999, a sensitivity of 95.3%, and a specificity of 98.7% for detecting small PDAC less than 2 cm in diameter, precisely localizing PDAC as small as 2 mm. In an external test involving 7,158 patients across 6 centers, ePAI achieved an AUC of 0.918-0.945, a sensitivity of 91.5%, and a specificity of 88.0%, precisely localizing PDAC as small as 5 mm. Importantly, ePAI detected PDACs on prediagnostic CT scans obtained 3 to 36 months before clinical diagnosis that had originally been overlooked by radiologists. It successfully detected and localized PDACs in 75 of 159 patients, with a median lead time of 347 days before clinical diagnosis. Our multi-reader study showed that ePAI significantly outperformed 30 board-certified radiologists by 50.3% (P < 0.05) in sensitivity while maintaining a comparable specificity of 95.4% in detecting PDACs early and prediagnostic. These findings suggest its potential of ePAI as an assistive tool to improve early detection of pancreatic cancer.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Abstract:Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
PanTS: The Pancreatic Tumor Segmentation Dataset
Jul 02, 2025Abstract:PanTS is a large-scale, multi-institutional dataset curated to advance research in pancreatic CT analysis. It contains 36,390 CT scans from 145 medical centers, with expert-validated, voxel-wise annotations of over 993,000 anatomical structures, covering pancreatic tumors, pancreas head, body, and tail, and 24 surrounding anatomical structures such as vascular/skeletal structures and abdominal/thoracic organs. Each scan includes metadata such as patient age, sex, diagnosis, contrast phase, in-plane spacing, slice thickness, etc. AI models trained on PanTS achieve significantly better performance in pancreatic tumor detection, localization, and segmentation compared to those trained on existing public datasets. Our analysis indicates that these gains are directly attributable to the 16x larger-scale tumor annotations and indirectly supported by the 24 additional surrounding anatomical structures. As the largest and most comprehensive resource of its kind, PanTS offers a new benchmark for developing and evaluating AI models in pancreatic CT analysis.
DSOcc: Leveraging Depth Awareness and Semantic Aid to Boost Camera-Based 3D Semantic Occupancy Prediction
May 27, 2025Abstract:Camera-based 3D semantic occupancy prediction offers an efficient and cost-effective solution for perceiving surrounding scenes in autonomous driving. However, existing works rely on explicit occupancy state inference, leading to numerous incorrect feature assignments, and insufficient samples restrict the learning of occupancy class inference. To address these challenges, we propose leveraging Depth awareness and Semantic aid to boost camera-based 3D semantic Occupancy prediction (DSOcc). We jointly perform occupancy state and occupancy class inference, where soft occupancy confidence is calculated through non-learning method and multiplied with image features to make the voxel representation aware of depth, enabling adaptive implicit occupancy state inference. Rather than focusing on improving feature learning, we directly utilize well-trained image semantic segmentation and fuse multiple frames with their occupancy probabilities to aid occupancy class inference, thereby enhancing robustness. Experimental results demonstrate that DSOcc achieves state-of-the-art performance on the SemanticKITTI dataset among camera-based methods.
Heterogeneous Data Game: Characterizing the Model Competition Across Multiple Data Sources
May 12, 2025Abstract:Data heterogeneity across multiple sources is common in real-world machine learning (ML) settings. Although many methods focus on enabling a single model to handle diverse data, real-world markets often comprise multiple competing ML providers. In this paper, we propose a game-theoretic framework -- the Heterogeneous Data Game -- to analyze how such providers compete across heterogeneous data sources. We investigate the resulting pure Nash equilibria (PNE), showing that they can be non-existent, homogeneous (all providers converge on the same model), or heterogeneous (providers specialize in distinct data sources). Our analysis spans monopolistic, duopolistic, and more general markets, illustrating how factors such as the "temperature" of data-source choice models and the dominance of certain data sources shape equilibrium outcomes. We offer theoretical insights into both homogeneous and heterogeneous PNEs, guiding regulatory policies and practical strategies for competitive ML marketplaces.
MedSegFactory: Text-Guided Generation of Medical Image-Mask Pairs
Apr 09, 2025Abstract:This paper presents MedSegFactory, a versatile medical synthesis framework that generates high-quality paired medical images and segmentation masks across modalities and tasks. It aims to serve as an unlimited data repository, supplying image-mask pairs to enhance existing segmentation tools. The core of MedSegFactory is a dual-stream diffusion model, where one stream synthesizes medical images and the other generates corresponding segmentation masks. To ensure precise alignment between image-mask pairs, we introduce Joint Cross-Attention (JCA), enabling a collaborative denoising paradigm by dynamic cross-conditioning between streams. This bidirectional interaction allows both representations to guide each other's generation, enhancing consistency between generated pairs. MedSegFactory unlocks on-demand generation of paired medical images and segmentation masks through user-defined prompts that specify the target labels, imaging modalities, anatomical regions, and pathological conditions, facilitating scalable and high-quality data generation. This new paradigm of medical image synthesis enables seamless integration into diverse medical imaging workflows, enhancing both efficiency and accuracy. Extensive experiments show that MedSegFactory generates data of superior quality and usability, achieving competitive or state-of-the-art performance in 2D and 3D segmentation tasks while addressing data scarcity and regulatory constraints.
RadGPT: Constructing 3D Image-Text Tumor Datasets
Jan 08, 2025Abstract:With over 85 million CT scans performed annually in the United States, creating tumor-related reports is a challenging and time-consuming task for radiologists. To address this need, we present RadGPT, an Anatomy-Aware Vision-Language AI Agent for generating detailed reports from CT scans. RadGPT first segments tumors, including benign cysts and malignant tumors, and their surrounding anatomical structures, then transforms this information into both structured reports and narrative reports. These reports provide tumor size, shape, location, attenuation, volume, and interactions with surrounding blood vessels and organs. Extensive evaluation on unseen hospitals shows that RadGPT can produce accurate reports, with high sensitivity/specificity for small tumor (<2 cm) detection: 80/73% for liver tumors, 92/78% for kidney tumors, and 77/77% for pancreatic tumors. For large tumors, sensitivity ranges from 89% to 97%. The results significantly surpass the state-of-the-art in abdominal CT report generation. RadGPT generated reports for 17 public datasets. Through radiologist review and refinement, we have ensured the reports' accuracy, and created the first publicly available image-text 3D medical dataset, comprising over 1.8 million text tokens and 2.7 million images from 9,262 CT scans, including 2,947 tumor scans/reports of 8,562 tumor instances. Our reports can: (1) localize tumors in eight liver sub-segments and three pancreatic sub-segments annotated per-voxel; (2) determine pancreatic tumor stage (T1-T4) in 260 reports; and (3) present individual analyses of multiple tumors--rare in human-made reports. Importantly, 948 of the reports are for early-stage tumors.
ScaleMAI: Accelerating the Development of Trusted Datasets and AI Models
Jan 06, 2025

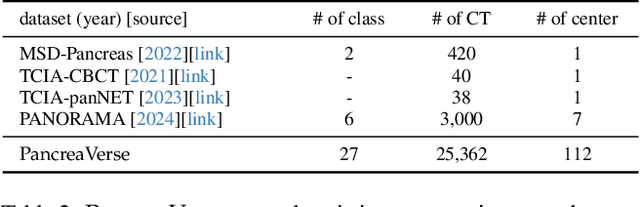

Abstract:Building trusted datasets is critical for transparent and responsible Medical AI (MAI) research, but creating even small, high-quality datasets can take years of effort from multidisciplinary teams. This process often delays AI benefits, as human-centric data creation and AI-centric model development are treated as separate, sequential steps. To overcome this, we propose ScaleMAI, an agent of AI-integrated data curation and annotation, allowing data quality and AI performance to improve in a self-reinforcing cycle and reducing development time from years to months. We adopt pancreatic tumor detection as an example. First, ScaleMAI progressively creates a dataset of 25,362 CT scans, including per-voxel annotations for benign/malignant tumors and 24 anatomical structures. Second, through progressive human-in-the-loop iterations, ScaleMAI provides Flagship AI Model that can approach the proficiency of expert annotators (30-year experience) in detecting pancreatic tumors. Flagship Model significantly outperforms models developed from smaller, fixed-quality datasets, with substantial gains in tumor detection (+14%), segmentation (+5%), and classification (72%) on three prestigious benchmarks. In summary, ScaleMAI transforms the speed, scale, and reliability of medical dataset creation, paving the way for a variety of impactful, data-driven applications.
Text-Driven Tumor Synthesis
Dec 24, 2024



Abstract:Tumor synthesis can generate examples that AI often misses or over-detects, improving AI performance by training on these challenging cases. However, existing synthesis methods, which are typically unconditional -- generating images from random variables -- or conditioned only by tumor shapes, lack controllability over specific tumor characteristics such as texture, heterogeneity, boundaries, and pathology type. As a result, the generated tumors may be overly similar or duplicates of existing training data, failing to effectively address AI's weaknesses. We propose a new text-driven tumor synthesis approach, termed TextoMorph, that provides textual control over tumor characteristics. This is particularly beneficial for examples that confuse the AI the most, such as early tumor detection (increasing Sensitivity by +8.5%), tumor segmentation for precise radiotherapy (increasing DSC by +6.3%), and classification between benign and malignant tumors (improving Sensitivity by +8.2%). By incorporating text mined from radiology reports into the synthesis process, we increase the variability and controllability of the synthetic tumors to target AI's failure cases more precisely. Moreover, TextoMorph uses contrastive learning across different texts and CT scans, significantly reducing dependence on scarce image-report pairs (only 141 pairs used in this study) by leveraging a large corpus of 34,035 radiology reports. Finally, we have developed rigorous tests to evaluate synthetic tumors, including Text-Driven Visual Turing Test and Radiomics Pattern Analysis, showing that our synthetic tumors is realistic and diverse in texture, heterogeneity, boundaries, and pathology.
Comprehensive Performance Evaluation of YOLOv11, YOLOv10, YOLOv9, YOLOv8 and YOLOv5 on Object Detection of Power Equipment
Nov 28, 2024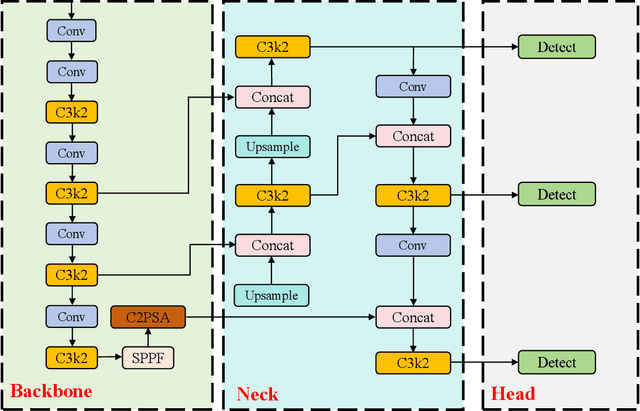


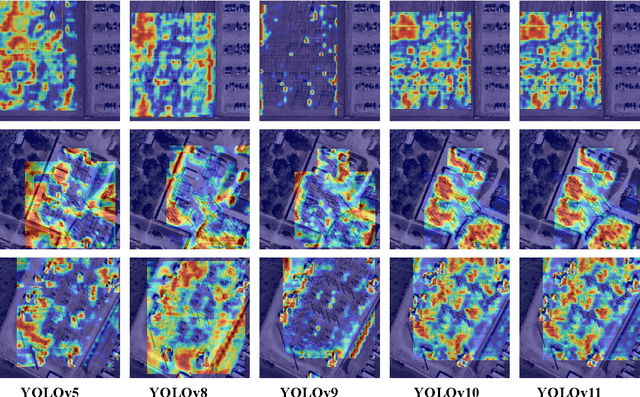
Abstract:With the rapid development of global industrial production, the demand for reliability in power equipment has been continuously increasing. Ensuring the stability of power system operations requires accurate methods to detect potential faults in power equipment, thereby guaranteeing the normal supply of electrical energy. In this article, the performance of YOLOv5, YOLOv8, YOLOv9, YOLOv10, and the state-of-the-art YOLOv11 methods was comprehensively evaluated for power equipment object detection. Experimental results demonstrate that the mean average precision (mAP) on a public dataset for power equipment was 54.4%, 55.5%, 43.8%, 48.0%, and 57.2%, respectively, with the YOLOv11 achieving the highest detection performance. Moreover, the YOLOv11 outperformed other methods in terms of recall rate and exhibited superior performance in reducing false detections. In conclusion, the findings indicate that the YOLOv11 model provides a reliable and effective solution for power equipment object detection, representing a promising approach to enhancing the operational reliability of power systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge