Haoliang Li
Nanyang Technological University, Singapore
Variation-Bounded Loss for Noise-Tolerant Learning
Nov 15, 2025Abstract:Mitigating the negative impact of noisy labels has been aperennial issue in supervised learning. Robust loss functions have emerged as a prevalent solution to this problem. In this work, we introduce the Variation Ratio as a novel property related to the robustness of loss functions, and propose a new family of robust loss functions, termed Variation-Bounded Loss (VBL), which is characterized by a bounded variation ratio. We provide theoretical analyses of the variation ratio, proving that a smaller variation ratio would lead to better robustness. Furthermore, we reveal that the variation ratio provides a feasible method to relax the symmetric condition and offers a more concise path to achieve the asymmetric condition. Based on the variation ratio, we reformulate several commonly used loss functions into a variation-bounded form for practical applications. Positive experiments on various datasets exhibit the effectiveness and flexibility of our approach.
Unveiling Modality Bias: Automated Sample-Specific Analysis for Multimodal Misinformation Benchmarks
Nov 08, 2025Abstract:Numerous multimodal misinformation benchmarks exhibit bias toward specific modalities, allowing detectors to make predictions based solely on one modality. While previous research has quantified bias at the dataset level or manually identified spurious correlations between modalities and labels, these approaches lack meaningful insights at the sample level and struggle to scale to the vast amount of online information. In this paper, we investigate the design for automated recognition of modality bias at the sample level. Specifically, we propose three bias quantification methods based on theories/views of different levels of granularity: 1) a coarse-grained evaluation of modality benefit; 2) a medium-grained quantification of information flow; and 3) a fine-grained causality analysis. To verify the effectiveness, we conduct a human evaluation on two popular benchmarks. Experimental results reveal three interesting findings that provide potential direction toward future research: 1)~Ensembling multiple views is crucial for reliable automated analysis; 2)~Automated analysis is prone to detector-induced fluctuations; and 3)~Different views produce a higher agreement on modality-balanced samples but diverge on biased ones.
Propose and Rectify: A Forensics-Driven MLLM Framework for Image Manipulation Localization
Aug 25, 2025Abstract:The increasing sophistication of image manipulation techniques demands robust forensic solutions that can both reliably detect alterations and precisely localize tampered regions. Recent Multimodal Large Language Models (MLLMs) show promise by leveraging world knowledge and semantic understanding for context-aware detection, yet they struggle with perceiving subtle, low-level forensic artifacts crucial for accurate manipulation localization. This paper presents a novel Propose-Rectify framework that effectively bridges semantic reasoning with forensic-specific analysis. In the proposal stage, our approach utilizes a forensic-adapted LLaVA model to generate initial manipulation analysis and preliminary localization of suspicious regions based on semantic understanding and contextual reasoning. In the rectification stage, we introduce a Forensics Rectification Module that systematically validates and refines these initial proposals through multi-scale forensic feature analysis, integrating technical evidence from several specialized filters. Additionally, we present an Enhanced Segmentation Module that incorporates critical forensic cues into SAM's encoded image embeddings, thereby overcoming inherent semantic biases to achieve precise delineation of manipulated regions. By synergistically combining advanced multimodal reasoning with established forensic methodologies, our framework ensures that initial semantic proposals are systematically validated and enhanced through concrete technical evidence, resulting in comprehensive detection accuracy and localization precision. Extensive experimental validation demonstrates state-of-the-art performance across diverse datasets with exceptional robustness and generalization capabilities.
Temporal Unlearnable Examples: Preventing Personal Video Data from Unauthorized Exploitation by Object Tracking
Jul 10, 2025Abstract:With the rise of social media, vast amounts of user-uploaded videos (e.g., YouTube) are utilized as training data for Visual Object Tracking (VOT). However, the VOT community has largely overlooked video data-privacy issues, as many private videos have been collected and used for training commercial models without authorization. To alleviate these issues, this paper presents the first investigation on preventing personal video data from unauthorized exploitation by deep trackers. Existing methods for preventing unauthorized data use primarily focus on image-based tasks (e.g., image classification), directly applying them to videos reveals several limitations, including inefficiency, limited effectiveness, and poor generalizability. To address these issues, we propose a novel generative framework for generating Temporal Unlearnable Examples (TUEs), and whose efficient computation makes it scalable for usage on large-scale video datasets. The trackers trained w/ TUEs heavily rely on unlearnable noises for temporal matching, ignoring the original data structure and thus ensuring training video data-privacy. To enhance the effectiveness of TUEs, we introduce a temporal contrastive loss, which further corrupts the learning of existing trackers when using our TUEs for training. Extensive experiments demonstrate that our approach achieves state-of-the-art performance in video data-privacy protection, with strong transferability across VOT models, datasets, and temporal matching tasks.
Disentangling Instruction Influence in Diffusion Transformers for Parallel Multi-Instruction-Guided Image Editing
Apr 07, 2025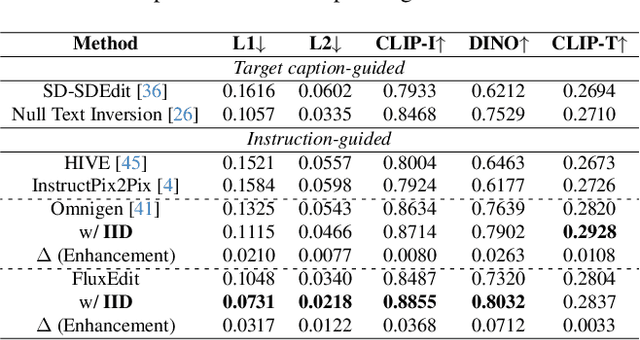
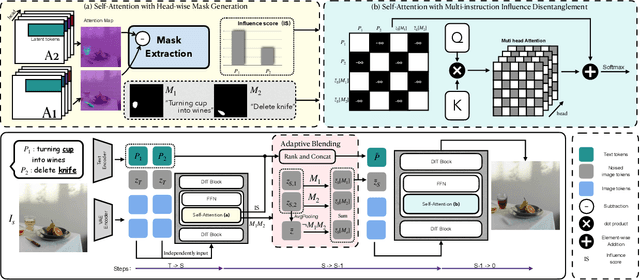
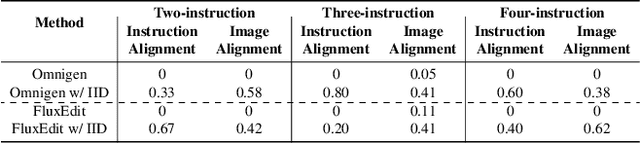
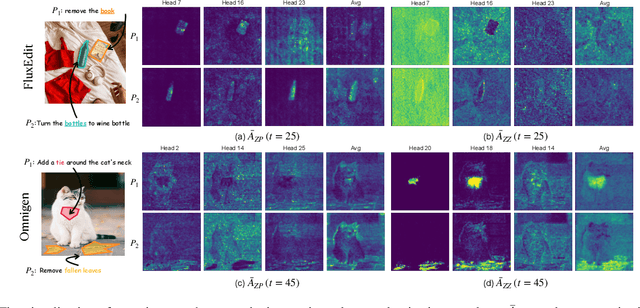
Abstract:Instruction-guided image editing enables users to specify modifications using natural language, offering more flexibility and control. Among existing frameworks, Diffusion Transformers (DiTs) outperform U-Net-based diffusion models in scalability and performance. However, while real-world scenarios often require concurrent execution of multiple instructions, step-by-step editing suffers from accumulated errors and degraded quality, and integrating multiple instructions with a single prompt usually results in incomplete edits due to instruction conflicts. We propose Instruction Influence Disentanglement (IID), a novel framework enabling parallel execution of multiple instructions in a single denoising process, designed for DiT-based models. By analyzing self-attention mechanisms in DiTs, we identify distinctive attention patterns in multi-instruction settings and derive instruction-specific attention masks to disentangle each instruction's influence. These masks guide the editing process to ensure localized modifications while preserving consistency in non-edited regions. Extensive experiments on open-source and custom datasets demonstrate that IID reduces diffusion steps while improving fidelity and instruction completion compared to existing baselines. The codes will be publicly released upon the acceptance of the paper.
SPACE: SPike-Aware Consistency Enhancement for Test-Time Adaptation in Spiking Neural Networks
Apr 03, 2025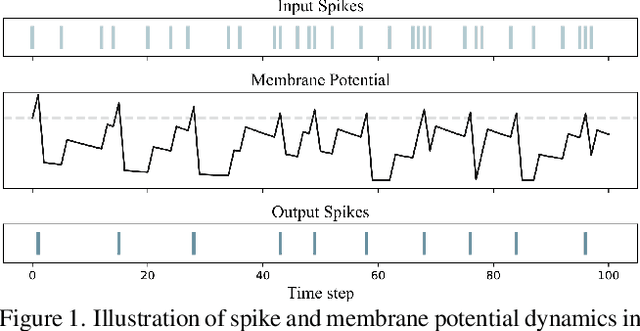

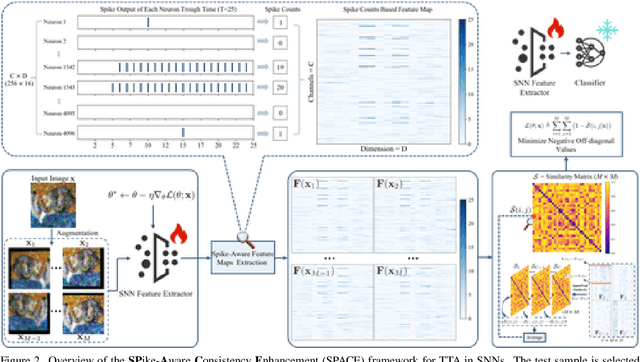

Abstract:Spiking Neural Networks (SNNs), as a biologically plausible alternative to Artificial Neural Networks (ANNs), have demonstrated advantages in terms of energy efficiency, temporal processing, and biological plausibility. However, SNNs are highly sensitive to distribution shifts, which can significantly degrade their performance in real-world scenarios. Traditional test-time adaptation (TTA) methods designed for ANNs often fail to address the unique computational dynamics of SNNs, such as sparsity and temporal spiking behavior. To address these challenges, we propose $\textbf{SP}$ike-$\textbf{A}$ware $\textbf{C}$onsistency $\textbf{E}$nhancement (SPACE), the first source-free and single-instance TTA method specifically designed for SNNs. SPACE leverages the inherent spike dynamics of SNNs to maximize the consistency of spike-behavior-based local feature maps across augmented versions of a single test sample, enabling robust adaptation without requiring source data. We evaluate SPACE on multiple datasets, including CIFAR-10-C, CIFAR-100-C, Tiny-ImageNet-C and DVS Gesture-C. Furthermore, SPACE demonstrates strong generalization across different model architectures, achieving consistent performance improvements on both VGG9 and ResNet11. Experimental results show that SPACE outperforms state-of-the-art methods, highlighting its effectiveness and robustness in real-world settings.
Test-time Adaptation for Foundation Medical Segmentation Model without Parametric Updates
Apr 02, 2025Abstract:Foundation medical segmentation models, with MedSAM being the most popular, have achieved promising performance across organs and lesions. However, MedSAM still suffers from compromised performance on specific lesions with intricate structures and appearance, as well as bounding box prompt-induced perturbations. Although current test-time adaptation (TTA) methods for medical image segmentation may tackle this issue, partial (e.g., batch normalization) or whole parametric updates restrict their effectiveness due to limited update signals or catastrophic forgetting in large models. Meanwhile, these approaches ignore the computational complexity during adaptation, which is particularly significant for modern foundation models. To this end, our theoretical analyses reveal that directly refining image embeddings is feasible to approach the same goal as parametric updates under the MedSAM architecture, which enables us to realize high computational efficiency and segmentation performance without the risk of catastrophic forgetting. Under this framework, we propose to encourage maximizing factorized conditional probabilities of the posterior prediction probability using a proposed distribution-approximated latent conditional random field loss combined with an entropy minimization loss. Experiments show that we achieve about 3\% Dice score improvements across three datasets while reducing computational complexity by over 7 times.
Enhancing Zero-Shot Image Recognition in Vision-Language Models through Human-like Concept Guidance
Mar 21, 2025Abstract:In zero-shot image recognition tasks, humans demonstrate remarkable flexibility in classifying unseen categories by composing known simpler concepts. However, existing vision-language models (VLMs), despite achieving significant progress through large-scale natural language supervision, often underperform in real-world applications because of sub-optimal prompt engineering and the inability to adapt effectively to target classes. To address these issues, we propose a Concept-guided Human-like Bayesian Reasoning (CHBR) framework. Grounded in Bayes' theorem, CHBR models the concept used in human image recognition as latent variables and formulates this task by summing across potential concepts, weighted by a prior distribution and a likelihood function. To tackle the intractable computation over an infinite concept space, we introduce an importance sampling algorithm that iteratively prompts large language models (LLMs) to generate discriminative concepts, emphasizing inter-class differences. We further propose three heuristic approaches involving Average Likelihood, Confidence Likelihood, and Test Time Augmentation (TTA) Likelihood, which dynamically refine the combination of concepts based on the test image. Extensive evaluations across fifteen datasets demonstrate that CHBR consistently outperforms existing state-of-the-art zero-shot generalization methods.
Q-PART: Quasi-Periodic Adaptive Regression with Test-time Training for Pediatric Left Ventricular Ejection Fraction Regression
Mar 06, 2025Abstract:In this work, we address the challenge of adaptive pediatric Left Ventricular Ejection Fraction (LVEF) assessment. While Test-time Training (TTT) approaches show promise for this task, they suffer from two significant limitations. Existing TTT works are primarily designed for classification tasks rather than continuous value regression, and they lack mechanisms to handle the quasi-periodic nature of cardiac signals. To tackle these issues, we propose a novel \textbf{Q}uasi-\textbf{P}eriodic \textbf{A}daptive \textbf{R}egression with \textbf{T}est-time Training (Q-PART) framework. In the training stage, the proposed Quasi-Period Network decomposes the echocardiogram into periodic and aperiodic components within latent space by combining parameterized helix trajectories with Neural Controlled Differential Equations. During inference, our framework further employs a variance minimization strategy across image augmentations that simulate common quality issues in echocardiogram acquisition, along with differential adaptation rates for periodic and aperiodic components. Theoretical analysis is provided to demonstrate that our variance minimization objective effectively bounds the regression error under mild conditions. Furthermore, extensive experiments across three pediatric age groups demonstrate that Q-PART not only significantly outperforms existing approaches in pediatric LVEF prediction, but also exhibits strong clinical screening capability with high mAUROC scores (up to 0.9747) and maintains gender-fair performance across all metrics, validating its robustness and practical utility in pediatric echocardiography analysis.
The NeRF Signature: Codebook-Aided Watermarking for Neural Radiance Fields
Feb 26, 2025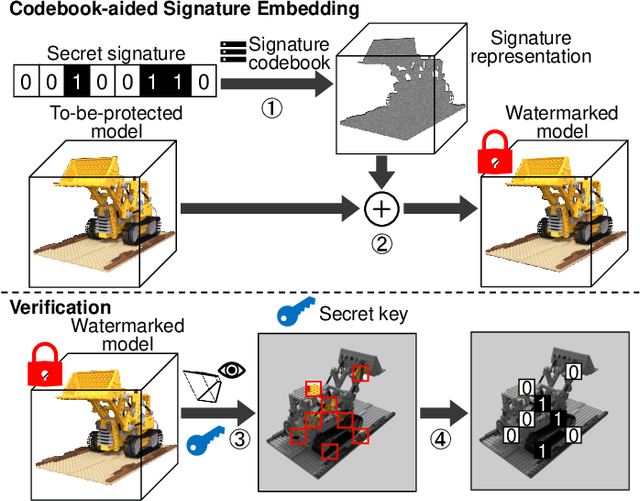
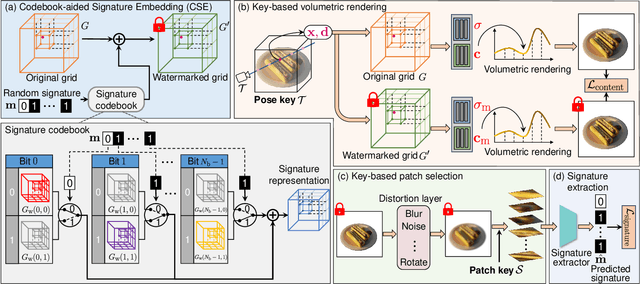
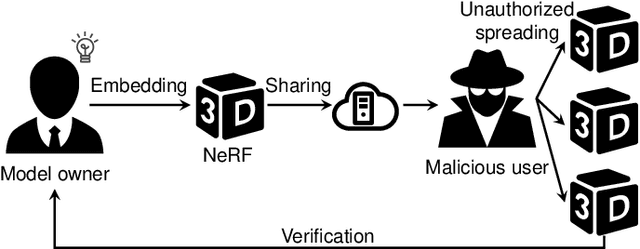
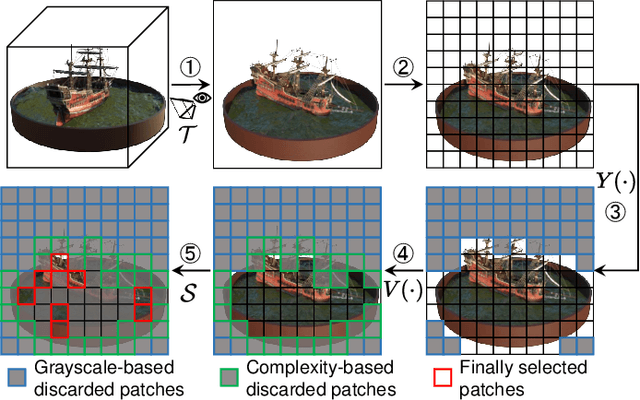
Abstract:Neural Radiance Fields (NeRF) have been gaining attention as a significant form of 3D content representation. With the proliferation of NeRF-based creations, the need for copyright protection has emerged as a critical issue. Although some approaches have been proposed to embed digital watermarks into NeRF, they often neglect essential model-level considerations and incur substantial time overheads, resulting in reduced imperceptibility and robustness, along with user inconvenience. In this paper, we extend the previous criteria for image watermarking to the model level and propose NeRF Signature, a novel watermarking method for NeRF. We employ a Codebook-aided Signature Embedding (CSE) that does not alter the model structure, thereby maintaining imperceptibility and enhancing robustness at the model level. Furthermore, after optimization, any desired signatures can be embedded through the CSE, and no fine-tuning is required when NeRF owners want to use new binary signatures. Then, we introduce a joint pose-patch encryption watermarking strategy to hide signatures into patches rendered from a specific viewpoint for higher robustness. In addition, we explore a Complexity-Aware Key Selection (CAKS) scheme to embed signatures in high visual complexity patches to enhance imperceptibility. The experimental results demonstrate that our method outperforms other baseline methods in terms of imperceptibility and robustness. The source code is available at: https://github.com/luo-ziyuan/NeRF_Signature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge