Pengfei Guo
SurgWorld: Learning Surgical Robot Policies from Videos via World Modeling
Dec 30, 2025Abstract:Data scarcity remains a fundamental barrier to achieving fully autonomous surgical robots. While large scale vision language action (VLA) models have shown impressive generalization in household and industrial manipulation by leveraging paired video action data from diverse domains, surgical robotics suffers from the paucity of datasets that include both visual observations and accurate robot kinematics. In contrast, vast corpora of surgical videos exist, but they lack corresponding action labels, preventing direct application of imitation learning or VLA training. In this work, we aim to alleviate this problem by learning policy models from SurgWorld, a world model designed for surgical physical AI. We curated the Surgical Action Text Alignment (SATA) dataset with detailed action description specifically for surgical robots. Then we built SurgeWorld based on the most advanced physical AI world model and SATA. It's able to generate diverse, generalizable and realistic surgery videos. We are also the first to use an inverse dynamics model to infer pseudokinematics from synthetic surgical videos, producing synthetic paired video action data. We demonstrate that a surgical VLA policy trained with these augmented data significantly outperforms models trained only on real demonstrations on a real surgical robot platform. Our approach offers a scalable path toward autonomous surgical skill acquisition by leveraging the abundance of unlabeled surgical video and generative world modeling, thus opening the door to generalizable and data efficient surgical robot policies.
SDUM: A Scalable Deep Unrolled Model for Universal MRI Reconstruction
Dec 19, 2025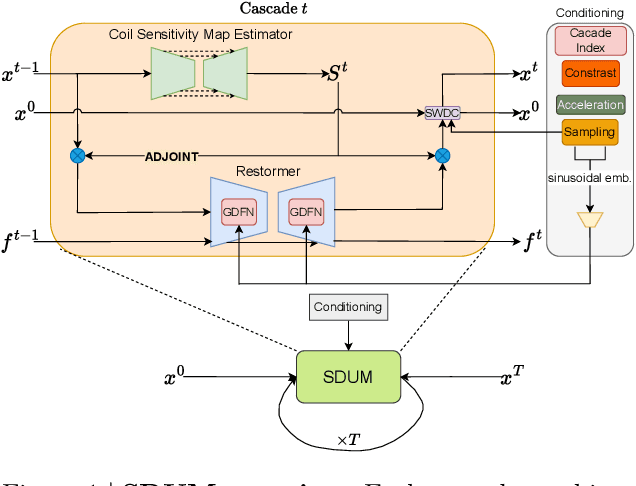
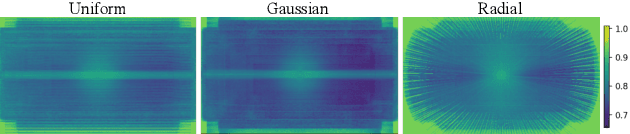

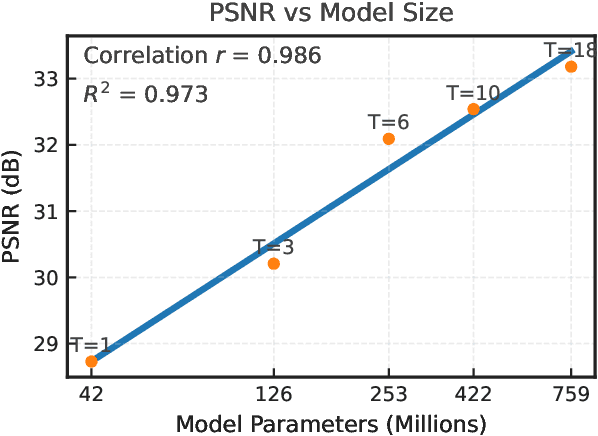
Abstract:Clinical MRI encompasses diverse imaging protocols--spanning anatomical targets (cardiac, brain, knee), contrasts (T1, T2, mapping), sampling patterns (Cartesian, radial, spiral, kt-space), and acceleration factors--yet current deep learning reconstructions are typically protocol-specific, hindering generalization and deployment. We introduce Scalable Deep Unrolled Model (SDUM), a universal framework combining a Restormer-based reconstructor, a learned coil sensitivity map estimator (CSME), sampling-aware weighted data consistency (SWDC), universal conditioning (UC) on cascade index and protocol metadata, and progressive cascade expansion training. SDUM exhibits foundation-model-like scaling behavior: reconstruction quality follows PSNR ${\sim}$ log(parameters) with correlation $r{=}0.986$ ($R^2{=}0.973$) up to 18 cascades, demonstrating predictable performance gains with model depth. A single SDUM trained on heterogeneous data achieves state-of-the-art results across all four CMRxRecon2025 challenge tracks--multi-center, multi-disease, 5T, and pediatric--without task-specific fine-tuning, surpassing specialized baselines by up to ${+}1.0$~dB. On CMRxRecon2024, SDUM outperforms the winning method PromptMR+ by ${+}0.55$~dB; on fastMRI brain, it exceeds PC-RNN by ${+}1.8$~dB. Ablations validate each component: SWDC ${+}0.43$~dB over standard DC, per-cascade CSME ${+}0.51$~dB, UC ${+}0.38$~dB. These results establish SDUM as a practical path toward universal, scalable MRI reconstruction.
Better Tokens for Better 3D: Advancing Vision-Language Modeling in 3D Medical Imaging
Oct 23, 2025Abstract:Recent progress in vision-language modeling for 3D medical imaging has been fueled by large-scale computed tomography (CT) corpora with paired free-text reports, stronger architectures, and powerful pretrained models. This has enabled applications such as automated report generation and text-conditioned 3D image synthesis. Yet, current approaches struggle with high-resolution, long-sequence volumes: contrastive pretraining often yields vision encoders that are misaligned with clinical language, and slice-wise tokenization blurs fine anatomy, reducing diagnostic performance on downstream tasks. We introduce BTB3D (Better Tokens for Better 3D), a causal convolutional encoder-decoder that unifies 2D and 3D training and inference while producing compact, frequency-aware volumetric tokens. A three-stage training curriculum enables (i) local reconstruction, (ii) overlapping-window tiling, and (iii) long-context decoder refinement, during which the model learns from short slice excerpts yet generalizes to scans exceeding 300 slices without additional memory overhead. BTB3D sets a new state-of-the-art on two key tasks: it improves BLEU scores and increases clinical F1 by 40% over CT2Rep, CT-CHAT, and Merlin for report generation; and it reduces FID by 75% and halves FVD compared to GenerateCT and MedSyn for text-to-CT synthesis, producing anatomically consistent 512*512*241 volumes. These results confirm that precise three-dimensional tokenization, rather than larger language backbones alone, is essential for scalable vision-language modeling in 3D medical imaging. The codebase is available at: https://github.com/ibrahimethemhamamci/BTB3D
Text2CT: Towards 3D CT Volume Generation from Free-text Descriptions Using Diffusion Model
May 07, 2025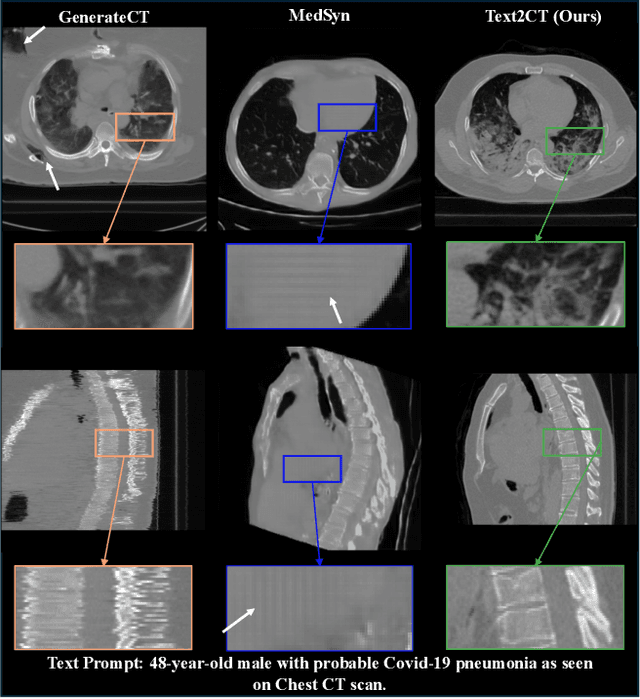
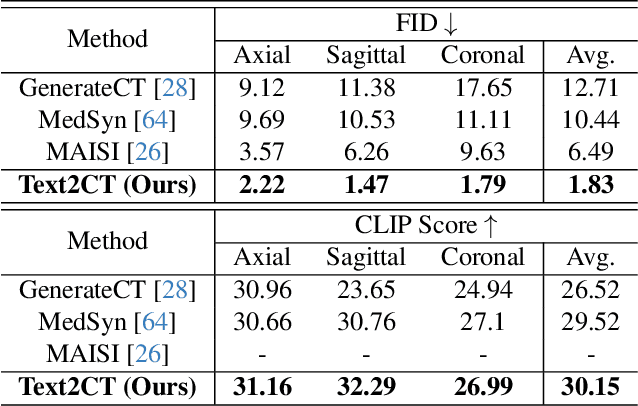
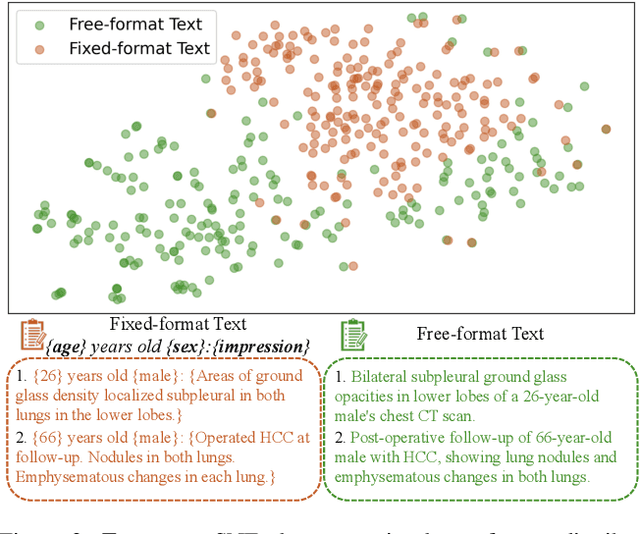
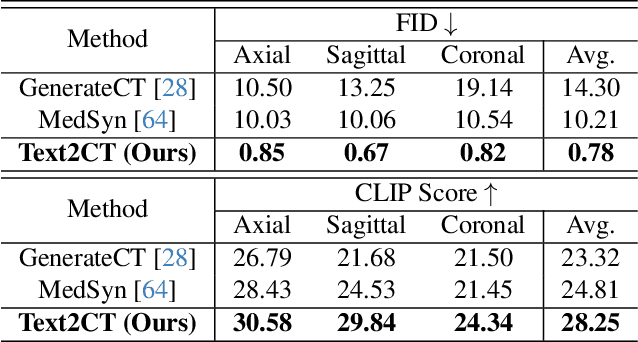
Abstract:Generating 3D CT volumes from descriptive free-text inputs presents a transformative opportunity in diagnostics and research. In this paper, we introduce Text2CT, a novel approach for synthesizing 3D CT volumes from textual descriptions using the diffusion model. Unlike previous methods that rely on fixed-format text input, Text2CT employs a novel prompt formulation that enables generation from diverse, free-text descriptions. The proposed framework encodes medical text into latent representations and decodes them into high-resolution 3D CT scans, effectively bridging the gap between semantic text inputs and detailed volumetric representations in a unified 3D framework. Our method demonstrates superior performance in preserving anatomical fidelity and capturing intricate structures as described in the input text. Extensive evaluations show that our approach achieves state-of-the-art results, offering promising potential applications in diagnostics, and data augmentation.
Text-Driven Tumor Synthesis
Dec 24, 2024



Abstract:Tumor synthesis can generate examples that AI often misses or over-detects, improving AI performance by training on these challenging cases. However, existing synthesis methods, which are typically unconditional -- generating images from random variables -- or conditioned only by tumor shapes, lack controllability over specific tumor characteristics such as texture, heterogeneity, boundaries, and pathology type. As a result, the generated tumors may be overly similar or duplicates of existing training data, failing to effectively address AI's weaknesses. We propose a new text-driven tumor synthesis approach, termed TextoMorph, that provides textual control over tumor characteristics. This is particularly beneficial for examples that confuse the AI the most, such as early tumor detection (increasing Sensitivity by +8.5%), tumor segmentation for precise radiotherapy (increasing DSC by +6.3%), and classification between benign and malignant tumors (improving Sensitivity by +8.2%). By incorporating text mined from radiology reports into the synthesis process, we increase the variability and controllability of the synthetic tumors to target AI's failure cases more precisely. Moreover, TextoMorph uses contrastive learning across different texts and CT scans, significantly reducing dependence on scarce image-report pairs (only 141 pairs used in this study) by leveraging a large corpus of 34,035 radiology reports. Finally, we have developed rigorous tests to evaluate synthetic tumors, including Text-Driven Visual Turing Test and Radiomics Pattern Analysis, showing that our synthetic tumors is realistic and diverse in texture, heterogeneity, boundaries, and pathology.
VILA-M3: Enhancing Vision-Language Models with Medical Expert Knowledge
Nov 19, 2024



Abstract:Generalist vision language models (VLMs) have made significant strides in computer vision, but they fall short in specialized fields like healthcare, where expert knowledge is essential. In traditional computer vision tasks, creative or approximate answers may be acceptable, but in healthcare, precision is paramount.Current large multimodal models like Gemini and GPT-4o are insufficient for medical tasks due to their reliance on memorized internet knowledge rather than the nuanced expertise required in healthcare. VLMs are usually trained in three stages: vision pre-training, vision-language pre-training, and instruction fine-tuning (IFT). IFT has been typically applied using a mixture of generic and healthcare data. In contrast, we propose that for medical VLMs, a fourth stage of specialized IFT is necessary, which focuses on medical data and includes information from domain expert models. Domain expert models developed for medical use are crucial because they are specifically trained for certain clinical tasks, e.g. to detect tumors and classify abnormalities through segmentation and classification, which learn fine-grained features of medical data$-$features that are often too intricate for a VLM to capture effectively especially in radiology. This paper introduces a new framework, VILA-M3, for medical VLMs that utilizes domain knowledge via expert models. Through our experiments, we show an improved state-of-the-art (SOTA) performance with an average improvement of ~9% over the prior SOTA model Med-Gemini and ~6% over models trained on the specific tasks. Our approach emphasizes the importance of domain expertise in creating precise, reliable VLMs for medical applications.
MAISI: Medical AI for Synthetic Imaging
Sep 13, 2024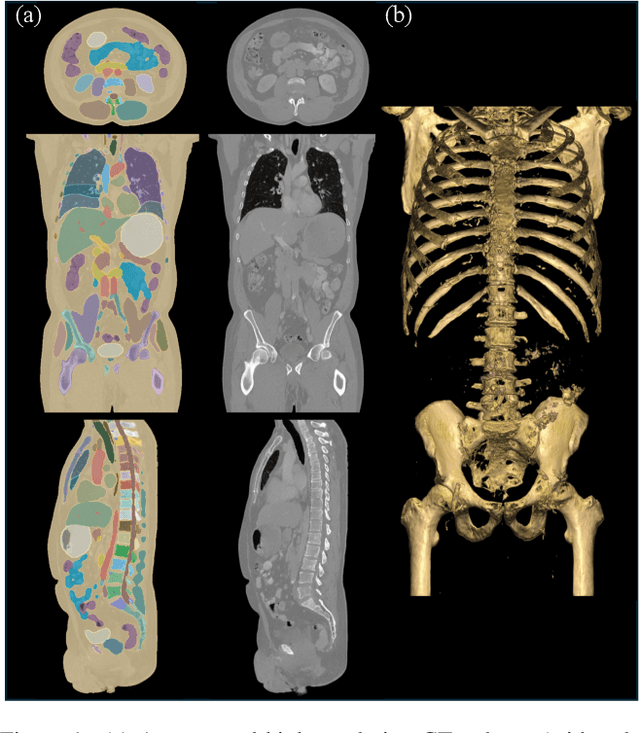
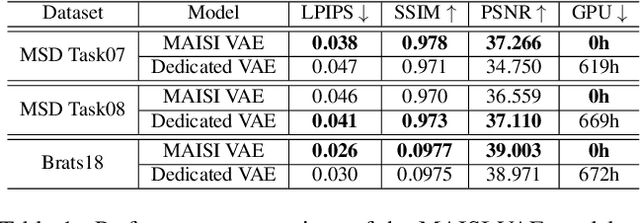
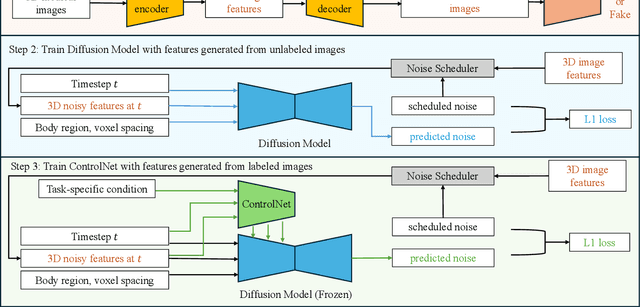
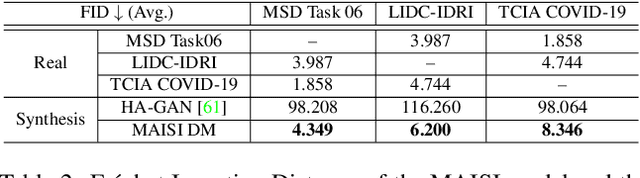
Abstract:Medical imaging analysis faces challenges such as data scarcity, high annotation costs, and privacy concerns. This paper introduces the Medical AI for Synthetic Imaging (MAISI), an innovative approach using the diffusion model to generate synthetic 3D computed tomography (CT) images to address those challenges. MAISI leverages the foundation volume compression network and the latent diffusion model to produce high-resolution CT images (up to a landmark volume dimension of 512 x 512 x 768 ) with flexible volume dimensions and voxel spacing. By incorporating ControlNet, MAISI can process organ segmentation, including 127 anatomical structures, as additional conditions and enables the generation of accurately annotated synthetic images that can be used for various downstream tasks. Our experiment results show that MAISI's capabilities in generating realistic, anatomically accurate images for diverse regions and conditions reveal its promising potential to mitigate challenges using synthetic data.
A Short Review and Evaluation of SAM2's Performance in 3D CT Image Segmentation
Aug 20, 2024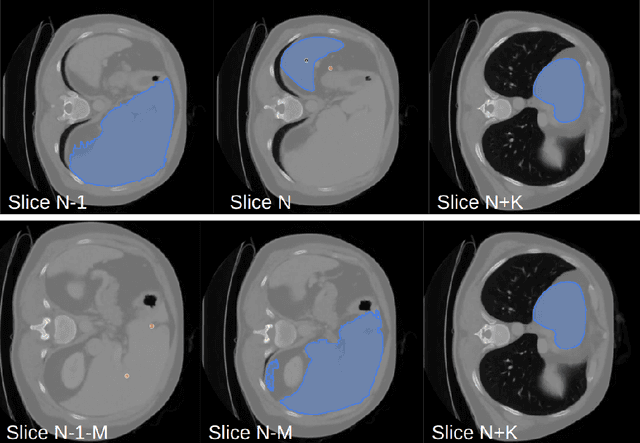
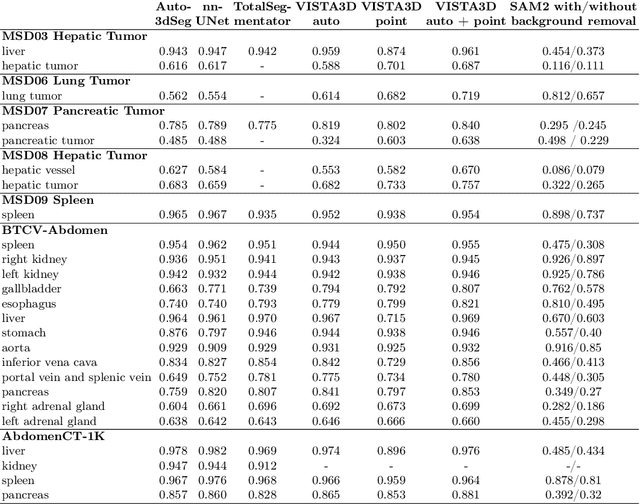
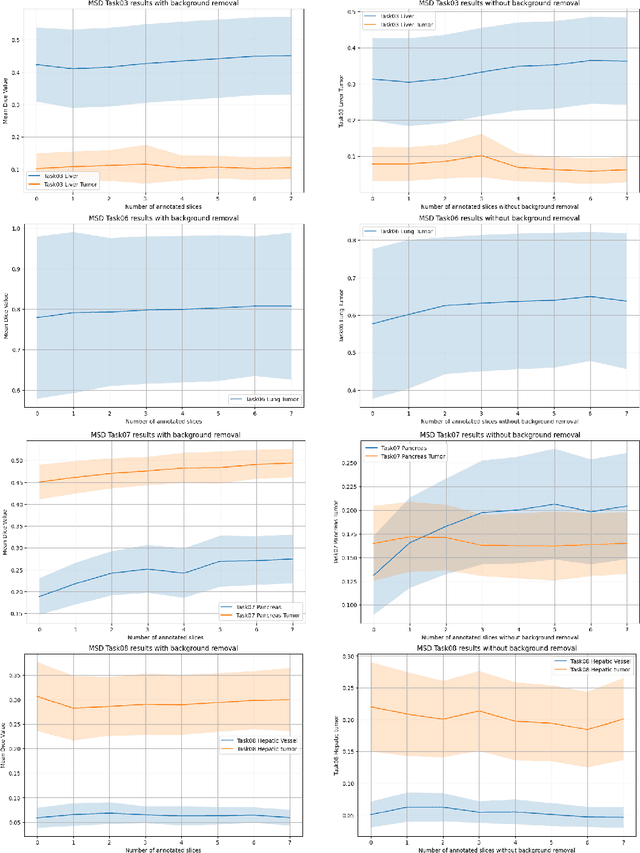
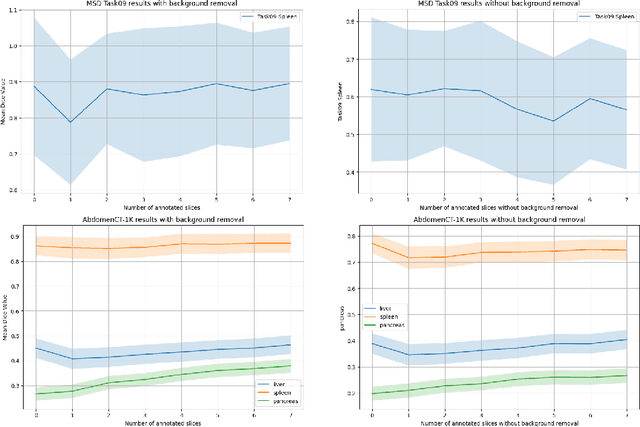
Abstract:Since the release of Segment Anything 2 (SAM2), the medical imaging community has been actively evaluating its performance for 3D medical image segmentation. However, different studies have employed varying evaluation pipelines, resulting in conflicting outcomes that obscure a clear understanding of SAM2's capabilities and potential applications. We shortly review existing benchmarks and point out that the SAM2 paper clearly outlines a zero-shot evaluation pipeline, which simulates user clicks iteratively for up to eight iterations. We reproduced this interactive annotation simulation on 3D CT datasets and provided the results and code~\url{https://github.com/Project-MONAI/VISTA}. Our findings reveal that directly applying SAM2 on 3D medical imaging in a zero-shot manner is far from satisfactory. It is prone to generating false positives when foreground objects disappear, and annotating more slices cannot fully offset this tendency. For smaller single-connected objects like kidney and aorta, SAM2 performs reasonably well but for most organs it is still far behind state-of-the-art 3D annotation methods. More research and innovation are needed for 3D medical imaging community to use SAM2 correctly.
VISTA3D: Versatile Imaging SegmenTation and Annotation model for 3D Computed Tomography
Jun 07, 2024Abstract:Segmentation foundation models have attracted great interest, however, none of them are adequate enough for the use cases in 3D computed tomography scans (CT) images. Existing works finetune on medical images with 2D foundation models trained on natural images, but interactive segmentation, especially in 2D, is too time-consuming for 3D scans and less useful for large cohort analysis. Models that can perform out-of-the-box automatic segmentation are more desirable. However, the model trained in this way lacks the ability to perform segmentation on unseen objects like novel tumors. Thus for 3D medical image analysis, an ideal segmentation solution might expect two features: accurate out-of-the-box performance covering major organ classes, and effective adaptation or zero-shot ability to novel structures. In this paper, we discuss what features a 3D CT segmentation foundation model should have, and introduce VISTA3D, Versatile Imaging SegmenTation and Annotation model. The model is trained systematically on 11454 volumes encompassing 127 types of human anatomical structures and various lesions and provides accurate out-of-the-box segmentation. The model's design also achieves state-of-the-art zero-shot interactive segmentation in 3D. The novel model design and training recipe represent a promising step toward developing a versatile medical image foundation model. Code and model weights will be released shortly. The early version of online demo can be tried on https://build.nvidia.com/nvidia/vista-3d.
Towards Federated Learning Under Resource Constraints via Layer-wise Training and Depth Dropout
Sep 11, 2023



Abstract:Large machine learning models trained on diverse data have recently seen unprecedented success. Federated learning enables training on private data that may otherwise be inaccessible, such as domain-specific datasets decentralized across many clients. However, federated learning can be difficult to scale to large models when clients have limited resources. This challenge often results in a trade-off between model size and access to diverse data. To mitigate this issue and facilitate training of large models on edge devices, we introduce a simple yet effective strategy, Federated Layer-wise Learning, to simultaneously reduce per-client memory, computation, and communication costs. Clients train just a single layer each round, reducing resource costs considerably with minimal performance degradation. We also introduce Federated Depth Dropout, a complementary technique that randomly drops frozen layers during training, to further reduce resource usage. Coupling these two techniques enables us to effectively train significantly larger models on edge devices. Specifically, we reduce training memory usage by 5x or more in federated self-supervised representation learning and demonstrate that performance in downstream tasks is comparable to conventional federated self-supervised learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge