Stephanie Harmon
DA-SSL: self-supervised domain adaptor to leverage foundational models in turbt histopathology slides
Dec 15, 2025Abstract:Recent deep learning frameworks in histopathology, particularly multiple instance learning (MIL) combined with pathology foundational models (PFMs), have shown strong performance. However, PFMs exhibit limitations on certain cancer or specimen types due to domain shifts - these cancer types were rarely used for pretraining or specimens contain tissue-based artifacts rarely seen within the pretraining population. Such is the case for transurethral resection of bladder tumor (TURBT), which are essential for diagnosing muscle-invasive bladder cancer (MIBC), but contain fragmented tissue chips and electrocautery artifacts and were not widely used in publicly available PFMs. To address this, we propose a simple yet effective domain-adaptive self-supervised adaptor (DA-SSL) that realigns pretrained PFM features to the TURBT domain without fine-tuning the foundational model itself. We pilot this framework for predicting treatment response in TURBT, where histomorphological features are currently underutilized and identifying patients who will benefit from neoadjuvant chemotherapy (NAC) is challenging. In our multi-center study, DA-SSL achieved an AUC of 0.77+/-0.04 in five-fold cross-validation and an external test accuracy of 0.84, sensitivity of 0.71, and specificity of 0.91 using majority voting. Our results demonstrate that lightweight domain adaptation with self-supervision can effectively enhance PFM-based MIL pipelines for clinically challenging histopathology tasks. Code is Available at https://github.com/zhanghaoyue/DA_SSL_TURBT.
MAISI: Medical AI for Synthetic Imaging
Sep 13, 2024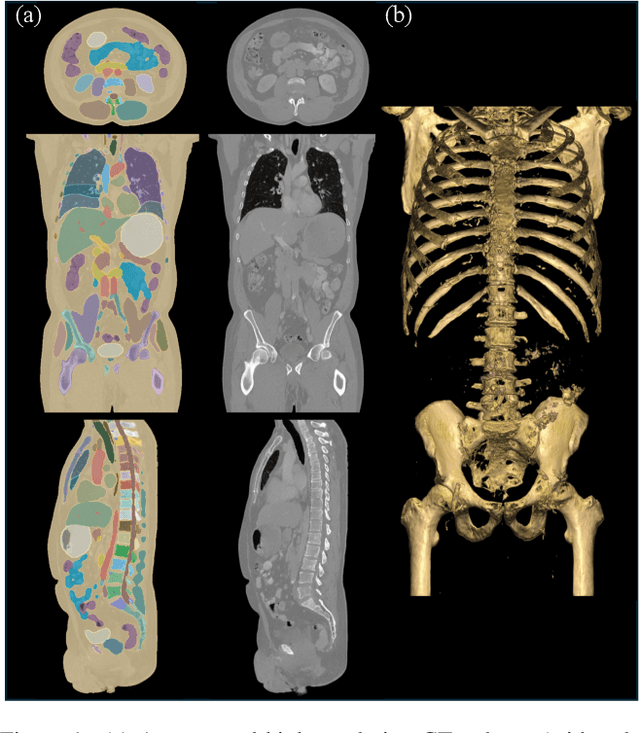
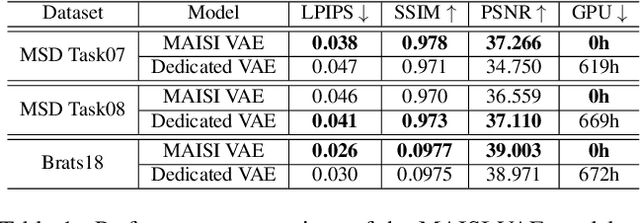
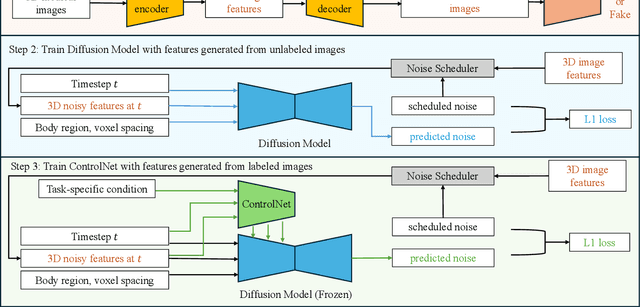
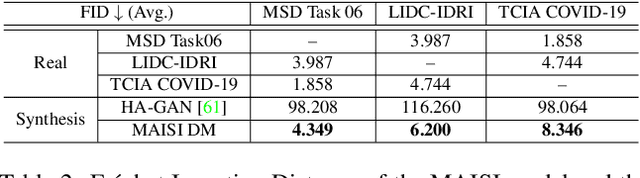
Abstract:Medical imaging analysis faces challenges such as data scarcity, high annotation costs, and privacy concerns. This paper introduces the Medical AI for Synthetic Imaging (MAISI), an innovative approach using the diffusion model to generate synthetic 3D computed tomography (CT) images to address those challenges. MAISI leverages the foundation volume compression network and the latent diffusion model to produce high-resolution CT images (up to a landmark volume dimension of 512 x 512 x 768 ) with flexible volume dimensions and voxel spacing. By incorporating ControlNet, MAISI can process organ segmentation, including 127 anatomical structures, as additional conditions and enables the generation of accurately annotated synthetic images that can be used for various downstream tasks. Our experiment results show that MAISI's capabilities in generating realistic, anatomically accurate images for diverse regions and conditions reveal its promising potential to mitigate challenges using synthetic data.
Location-based Radiology Report-Guided Semi-supervised Learning for Prostate Cancer Detection
Jun 18, 2024Abstract:Prostate cancer is one of the most prevalent malignancies in the world. While deep learning has potential to further improve computer-aided prostate cancer detection on MRI, its efficacy hinges on the exhaustive curation of manually annotated images. We propose a novel methodology of semisupervised learning (SSL) guided by automatically extracted clinical information, specifically the lesion locations in radiology reports, allowing for use of unannotated images to reduce the annotation burden. By leveraging lesion locations, we refined pseudo labels, which were then used to train our location-based SSL model. We show that our SSL method can improve prostate lesion detection by utilizing unannotated images, with more substantial impacts being observed when larger proportions of unannotated images are used.
VISTA3D: Versatile Imaging SegmenTation and Annotation model for 3D Computed Tomography
Jun 07, 2024Abstract:Segmentation foundation models have attracted great interest, however, none of them are adequate enough for the use cases in 3D computed tomography scans (CT) images. Existing works finetune on medical images with 2D foundation models trained on natural images, but interactive segmentation, especially in 2D, is too time-consuming for 3D scans and less useful for large cohort analysis. Models that can perform out-of-the-box automatic segmentation are more desirable. However, the model trained in this way lacks the ability to perform segmentation on unseen objects like novel tumors. Thus for 3D medical image analysis, an ideal segmentation solution might expect two features: accurate out-of-the-box performance covering major organ classes, and effective adaptation or zero-shot ability to novel structures. In this paper, we discuss what features a 3D CT segmentation foundation model should have, and introduce VISTA3D, Versatile Imaging SegmenTation and Annotation model. The model is trained systematically on 11454 volumes encompassing 127 types of human anatomical structures and various lesions and provides accurate out-of-the-box segmentation. The model's design also achieves state-of-the-art zero-shot interactive segmentation in 3D. The novel model design and training recipe represent a promising step toward developing a versatile medical image foundation model. Code and model weights will be released shortly. The early version of online demo can be tried on https://build.nvidia.com/nvidia/vista-3d.
Using YOLO v7 to Detect Kidney in Magnetic Resonance Imaging
Feb 12, 2024Abstract:Introduction This study explores the use of the latest You Only Look Once (YOLO V7) object detection method to enhance kidney detection in medical imaging by training and testing a modified YOLO V7 on medical image formats. Methods Study includes 878 patients with various subtypes of renal cell carcinoma (RCC) and 206 patients with normal kidneys. A total of 5657 MRI scans for 1084 patients were retrieved. 326 patients with 1034 tumors recruited from a retrospective maintained database, and bounding boxes were drawn around their tumors. A primary model was trained on 80% of annotated cases, with 20% saved for testing (primary test set). The best primary model was then used to identify tumors in the remaining 861 patients and bounding box coordinates were generated on their scans using the model. Ten benchmark training sets were created with generated coordinates on not-segmented patients. The final model used to predict the kidney in the primary test set. We reported the positive predictive value (PPV), sensitivity, and mean average precision (mAP). Results The primary training set showed an average PPV of 0.94 +/- 0.01, sensitivity of 0.87 +/- 0.04, and mAP of 0.91 +/- 0.02. The best primary model yielded a PPV of 0.97, sensitivity of 0.92, and mAP of 0.95. The final model demonstrated an average PPV of 0.95 +/- 0.03, sensitivity of 0.98 +/- 0.004, and mAP of 0.95 +/- 0.01. Conclusion Using a semi-supervised approach with a medical image library, we developed a high-performing model for kidney detection. Further external validation is required to assess the model's generalizability.
Auto-FedRL: Federated Hyperparameter Optimization for Multi-institutional Medical Image Segmentation
Mar 12, 2022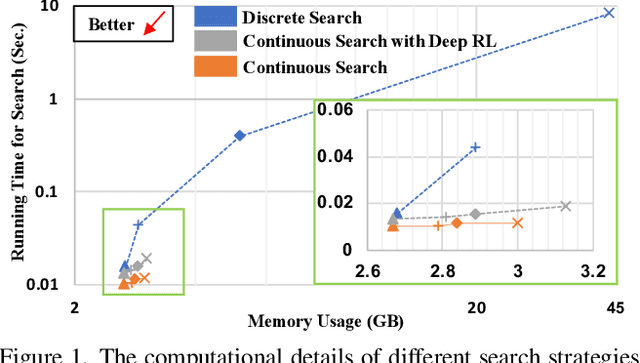
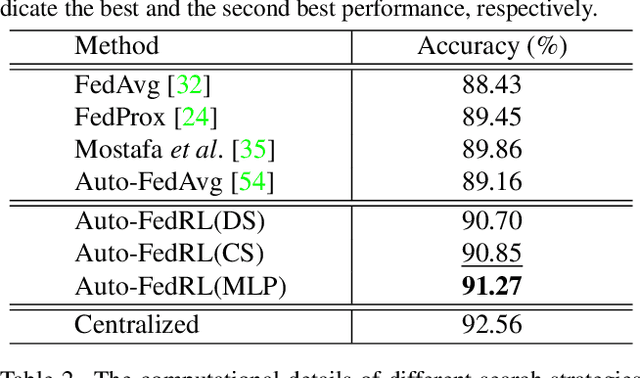
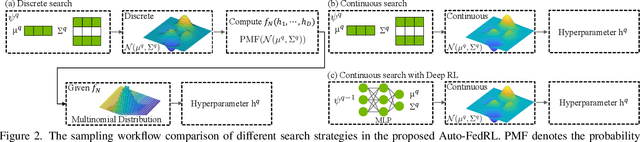

Abstract:Federated learning (FL) is a distributed machine learning technique that enables collaborative model training while avoiding explicit data sharing. The inherent privacy-preserving property of FL algorithms makes them especially attractive to the medical field. However, in case of heterogeneous client data distributions, standard FL methods are unstable and require intensive hyperparameter tuning to achieve optimal performance. Conventional hyperparameter optimization algorithms are impractical in real-world FL applications as they involve numerous training trials, which are often not affordable with limited compute budgets. In this work, we propose an efficient reinforcement learning~(RL)-based federated hyperparameter optimization algorithm, termed Auto-FedRL, in which an online RL agent can dynamically adjust hyperparameters of each client based on the current training progress. Extensive experiments are conducted to investigate different search strategies and RL agents. The effectiveness of the proposed method is validated on a heterogeneous data split of the CIFAR-10 dataset as well as two real-world medical image segmentation datasets for COVID-19 lesion segmentation in chest CT and pancreas segmentation in abdominal CT.
Auto-FedAvg: Learnable Federated Averaging for Multi-Institutional Medical Image Segmentation
Apr 20, 2021



Abstract:Federated learning (FL) enables collaborative model training while preserving each participant's privacy, which is particularly beneficial to the medical field. FedAvg is a standard algorithm that uses fixed weights, often originating from the dataset sizes at each client, to aggregate the distributed learned models on a server during the FL process. However, non-identical data distribution across clients, known as the non-i.i.d problem in FL, could make this assumption for setting fixed aggregation weights sub-optimal. In this work, we design a new data-driven approach, namely Auto-FedAvg, where aggregation weights are dynamically adjusted, depending on data distributions across data silos and the current training progress of the models. We disentangle the parameter set into two parts, local model parameters and global aggregation parameters, and update them iteratively with a communication-efficient algorithm. We first show the validity of our approach by outperforming state-of-the-art FL methods for image recognition on a heterogeneous data split of CIFAR-10. Furthermore, we demonstrate our algorithm's effectiveness on two multi-institutional medical image analysis tasks, i.e., COVID-19 lesion segmentation in chest CT and pancreas segmentation in abdominal CT.
Federated Semi-Supervised Learning for COVID Region Segmentation in Chest CT using Multi-National Data from China, Italy, Japan
Nov 23, 2020



Abstract:The recent outbreak of COVID-19 has led to urgent needs for reliable diagnosis and management of SARS-CoV-2 infection. As a complimentary tool, chest CT has been shown to be able to reveal visual patterns characteristic for COVID-19, which has definite value at several stages during the disease course. To facilitate CT analysis, recent efforts have focused on computer-aided characterization and diagnosis, which has shown promising results. However, domain shift of data across clinical data centers poses a serious challenge when deploying learning-based models. In this work, we attempt to find a solution for this challenge via federated and semi-supervised learning. A multi-national database consisting of 1704 scans from three countries is adopted to study the performance gap, when training a model with one dataset and applying it to another. Expert radiologists manually delineated 945 scans for COVID-19 findings. In handling the variability in both the data and annotations, a novel federated semi-supervised learning technique is proposed to fully utilize all available data (with or without annotations). Federated learning avoids the need for sensitive data-sharing, which makes it favorable for institutions and nations with strict regulatory policy on data privacy. Moreover, semi-supervision potentially reduces the annotation burden under a distributed setting. The proposed framework is shown to be effective compared to fully supervised scenarios with conventional data sharing instead of model weight sharing.
When Unseen Domain Generalization is Unnecessary? Rethinking Data Augmentation
Jun 12, 2019



Abstract:Recent advances in deep learning for medical image segmentation demonstrate expert-level accuracy. However, in clinically realistic environments, such methods have marginal performance due to differences in image domains, including different imaging protocols, device vendors and patient populations. Here we consider the problem of domain generalization, when a model is trained once, and its performance generalizes to unseen domains. Intuitively, within a specific medical imaging modality the domain differences are smaller relative to natural images domain variability. We rethink data augmentation for medical 3D images and propose a deep stacked transformations (DST) approach for domain generalization. Specifically, a series of n stacked transformations are applied to each image in each mini-batch during network training to account for the contribution of domain-specific shifts in medical images. We comprehensively evaluate our method on three tasks: segmentation of whole prostate from 3D MRI, left atrial from 3D MRI, and left ventricle from 3D ultrasound. We demonstrate that when trained on a small source dataset, (i) on average, DST models on unseen datasets degrade only by 11% (Dice score change), compared to the conventional augmentation (degrading 39%) and CycleGAN-based domain adaptation method (degrading 25%); (ii) when evaluation on the same domain, DST is also better albeit only marginally. (iii) When training on large-sized data, DST on unseen domains reaches performance of state-of-the-art fully supervised models. These findings establish a strong benchmark for the study of domain generalization in medical imaging, and can be generalized to the design of robust deep segmentation models for clinical deployment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge