Ling Zhang
Missouri S&T EMC Laboratory, Rolla, MO, USA
Non-Contrast CT Esophageal Varices Grading through Clinical Prior-Enhanced Multi-Organ Analysis
Dec 22, 2025Abstract:Esophageal varices (EV) represent a critical complication of portal hypertension, affecting approximately 60% of cirrhosis patients with a significant bleeding risk of ~30%. While traditionally diagnosed through invasive endoscopy, non-contrast computed tomography (NCCT) presents a potential non-invasive alternative that has yet to be fully utilized in clinical practice. We present Multi-Organ-COhesion Network++ (MOON++), a novel multimodal framework that enhances EV assessment through comprehensive analysis of NCCT scans. Inspired by clinical evidence correlating organ volumetric relationships with liver disease severity, MOON++ synthesizes imaging characteristics of the esophagus, liver, and spleen through multimodal learning. We evaluated our approach using 1,631 patients, those with endoscopically confirmed EV were classified into four severity grades. Validation in 239 patient cases and independent testing in 289 cases demonstrate superior performance compared to conventional single organ methods, achieving an AUC of 0.894 versus 0.803 for the severe grade EV classification (G3 versus <G3) and 0.921 versus 0.793 for the differentiation of moderate to severe grades (>=G2 versus <G2). We conducted a reader study involving experienced radiologists to further validate the performance of MOON++. To our knowledge, MOON++ represents the first comprehensive multi-organ NCCT analysis framework incorporating clinical knowledge priors for EV assessment, potentially offering a promising non-invasive diagnostic alternative.
Holdout-Loss-Based Data Selection for LLM Finetuning via In-Context Learning
Oct 16, 2025Abstract:Fine-tuning large pretrained language models is a common approach for aligning them with human preferences, but noisy or off-target examples can dilute supervision. While small, well-chosen datasets often match the performance of much larger ones, systematic and efficient ways to identify high-value training data remain underexplored. Many current methods rely on heuristics or expensive retraining. We present a theoretically grounded, resource-efficient framework for data selection and reweighting. At its core is an In-Context Approximation (ICA) that estimates the holdout loss a model would incur after training on a candidate example by conditioning on a small, curated holdout set in context. ICA requires no reference model and no additional finetuning. Under a local linearization, ICA is equivalent to a first-order update toward the holdout optimum, motivating its use as a proxy for data value. We derive per-example weights from ICA scores, dynamically reweighting gradient updates as model parameters evolve. Across SFT, DPO, and SimPO, and over diverse backbones and datasets, ICA-based reweighting consistently improves model alignment with minimal overhead. We analyze sensitivity to score update frequency and the choice of $k$ holdout examples for in-context demonstrations, and note limitations for rapidly drifting on-policy updates, highlighting directions for future work. Code and prompts will be released.
HeurAgenix: Leveraging LLMs for Solving Complex Combinatorial Optimization Challenges
Jun 18, 2025Abstract:Heuristic algorithms play a vital role in solving combinatorial optimization (CO) problems, yet traditional designs depend heavily on manual expertise and struggle to generalize across diverse instances. We introduce \textbf{HeurAgenix}, a two-stage hyper-heuristic framework powered by large language models (LLMs) that first evolves heuristics and then selects among them automatically. In the heuristic evolution phase, HeurAgenix leverages an LLM to compare seed heuristic solutions with higher-quality solutions and extract reusable evolution strategies. During problem solving, it dynamically picks the most promising heuristic for each problem state, guided by the LLM's perception ability. For flexibility, this selector can be either a state-of-the-art LLM or a fine-tuned lightweight model with lower inference cost. To mitigate the scarcity of reliable supervision caused by CO complexity, we fine-tune the lightweight heuristic selector with a dual-reward mechanism that jointly exploits singals from selection preferences and state perception, enabling robust selection under noisy annotations. Extensive experiments on canonical benchmarks show that HeurAgenix not only outperforms existing LLM-based hyper-heuristics but also matches or exceeds specialized solvers. Code is available at https://github.com/microsoft/HeurAgenix.
Leveraging Interview-Informed LLMs to Model Survey Responses: Comparative Insights from AI-Generated and Human Data
May 28, 2025Abstract:Mixed methods research integrates quantitative and qualitative data but faces challenges in aligning their distinct structures, particularly in examining measurement characteristics and individual response patterns. Advances in large language models (LLMs) offer promising solutions by generating synthetic survey responses informed by qualitative data. This study investigates whether LLMs, guided by personal interviews, can reliably predict human survey responses, using the Behavioral Regulations in Exercise Questionnaire (BREQ) and interviews from after-school program staff as a case study. Results indicate that LLMs capture overall response patterns but exhibit lower variability than humans. Incorporating interview data improves response diversity for some models (e.g., Claude, GPT), while well-crafted prompts and low-temperature settings enhance alignment between LLM and human responses. Demographic information had less impact than interview content on alignment accuracy. These findings underscore the potential of interview-informed LLMs to bridge qualitative and quantitative methodologies while revealing limitations in response variability, emotional interpretation, and psychometric fidelity. Future research should refine prompt design, explore bias mitigation, and optimize model settings to enhance the validity of LLM-generated survey data in social science research.
Large-scale and Fine-grained Vision-language Pre-training for Enhanced CT Image Understanding
Jan 24, 2025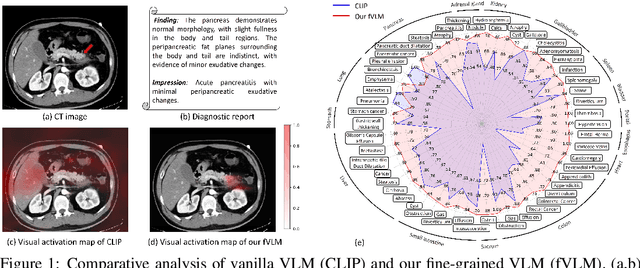
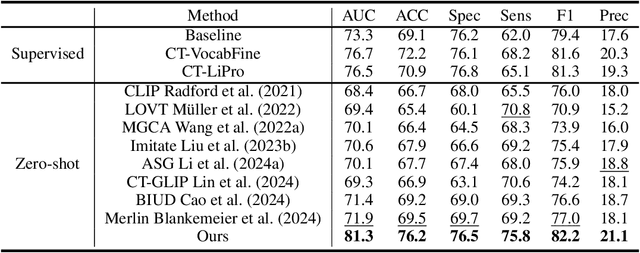
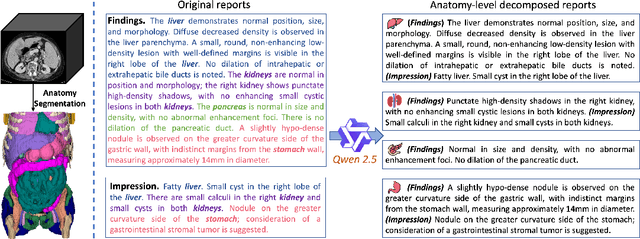
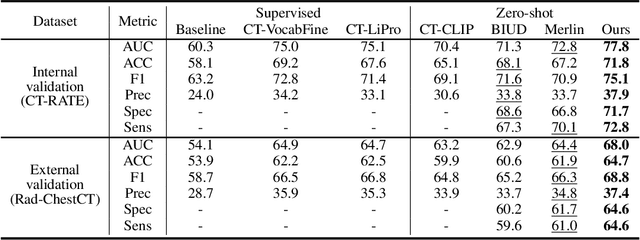
Abstract:Artificial intelligence (AI) shows great potential in assisting radiologists to improve the efficiency and accuracy of medical image interpretation and diagnosis. However, a versatile AI model requires large-scale data and comprehensive annotations, which are often impractical in medical settings. Recent studies leverage radiology reports as a naturally high-quality supervision for medical images, using contrastive language-image pre-training (CLIP) to develop language-informed models for radiological image interpretation. Nonetheless, these approaches typically contrast entire images with reports, neglecting the local associations between imaging regions and report sentences, which may undermine model performance and interoperability. In this paper, we propose a fine-grained vision-language model (fVLM) for anatomy-level CT image interpretation. Specifically, we explicitly match anatomical regions of CT images with corresponding descriptions in radiology reports and perform contrastive pre-training for each anatomy individually. Fine-grained alignment, however, faces considerable false-negative challenges, mainly from the abundance of anatomy-level healthy samples and similarly diseased abnormalities. To tackle this issue, we propose identifying false negatives of both normal and abnormal samples and calibrating contrastive learning from patient-level to disease-aware pairing. We curated the largest CT dataset to date, comprising imaging and report data from 69,086 patients, and conducted a comprehensive evaluation of 54 major and important disease diagnosis tasks across 15 main anatomies. Experimental results demonstrate the substantial potential of fVLM in versatile medical image interpretation. In the zero-shot classification task, we achieved an average AUC of 81.3% on 54 diagnosis tasks, surpassing CLIP and supervised methods by 12.9% and 8.0%, respectively.
Doc-Guided Sent2Sent++: A Sent2Sent++ Agent with Doc-Guided memory for Document-level Machine Translation
Jan 15, 2025



Abstract:The field of artificial intelligence has witnessed significant advancements in natural language processing, largely attributed to the capabilities of Large Language Models (LLMs). These models form the backbone of Agents designed to address long-context dependencies, particularly in Document-level Machine Translation (DocMT). DocMT presents unique challenges, with quality, consistency, and fluency being the key metrics for evaluation. Existing approaches, such as Doc2Doc and Doc2Sent, either omit sentences or compromise fluency. This paper introduces Doc-Guided Sent2Sent++, an Agent that employs an incremental sentence-level forced decoding strategy \textbf{to ensure every sentence is translated while enhancing the fluency of adjacent sentences.} Our Agent leverages a Doc-Guided Memory, focusing solely on the summary and its translation, which we find to be an efficient approach to maintaining consistency. Through extensive testing across multiple languages and domains, we demonstrate that Sent2Sent++ outperforms other methods in terms of quality, consistency, and fluency. The results indicate that, our approach has achieved significant improvements in metrics such as s-COMET, d-COMET, LTCR-$1_f$, and document-level perplexity (d-ppl). The contributions of this paper include a detailed analysis of current DocMT research, the introduction of the Sent2Sent++ decoding method, the Doc-Guided Memory mechanism, and validation of its effectiveness across languages and domains.
Post-hoc Interpretability Illumination for Scientific Interaction Discovery
Dec 20, 2024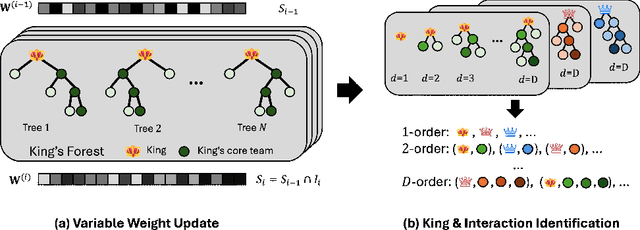

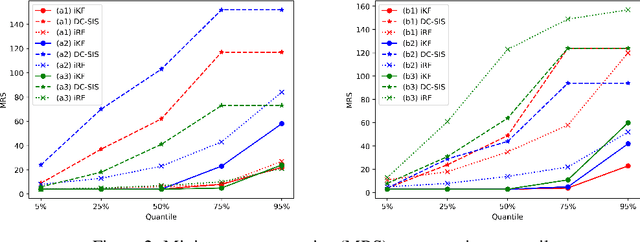
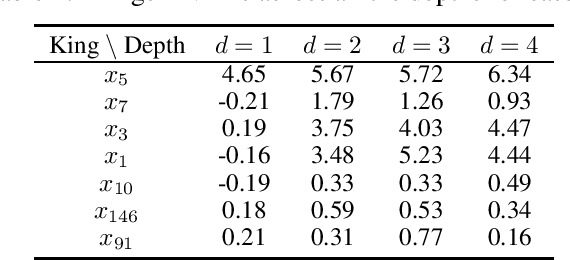
Abstract:Model interpretability and explainability have garnered substantial attention in recent years, particularly in decision-making applications. However, existing interpretability tools often fall short in delivering satisfactory performance due to limited capabilities or efficiency issues. To address these challenges, we propose a novel post-hoc method: Iterative Kings' Forests (iKF), designed to uncover complex multi-order interactions among variables. iKF iteratively selects the next most important variable, the "King", and constructs King's Forests by placing it at the root node of each tree to identify variables that interact with the "King". It then generates ranked short lists of important variables and interactions of varying orders. Additionally, iKF provides inference metrics to analyze the patterns of the selected interactions and classify them into one of three interaction types: Accompanied Interaction, Synergistic Interaction, and Hierarchical Interaction. Extensive experiments demonstrate the strong interpretive power of our proposed iKF, highlighting its great potential for explainable modeling and scientific discovery across diverse scientific fields.
Improved Esophageal Varices Assessment from Non-Contrast CT Scans
Jul 18, 2024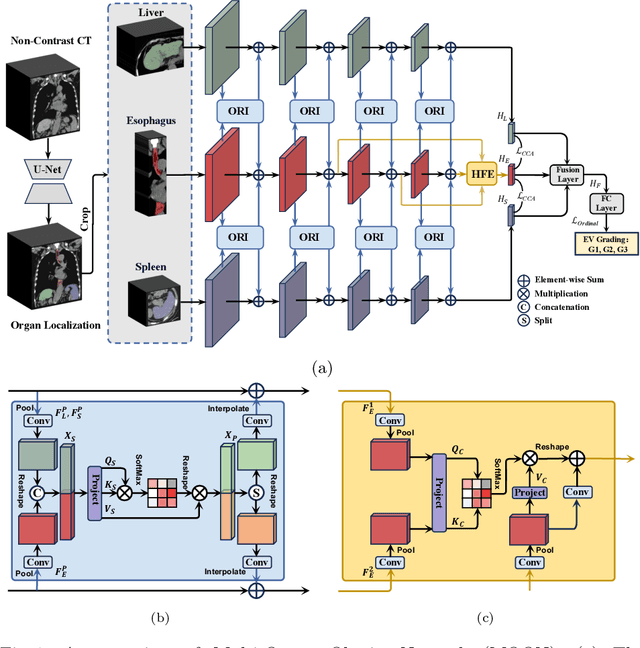
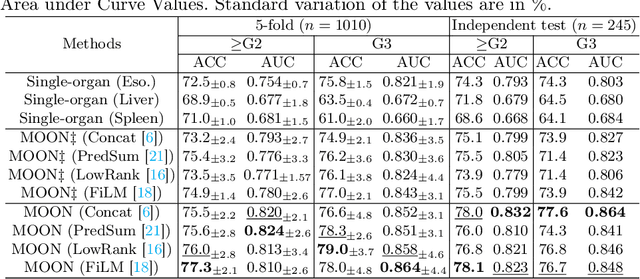
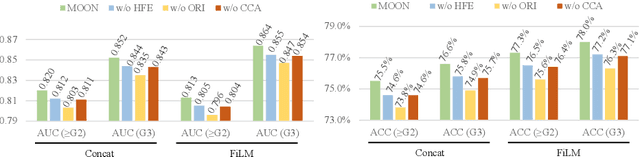
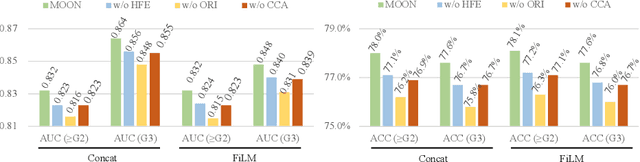
Abstract:Esophageal varices (EV), a serious health concern resulting from portal hypertension, are traditionally diagnosed through invasive endoscopic procedures. Despite non-contrast computed tomography (NC-CT) imaging being a less expensive and non-invasive imaging modality, it has yet to gain full acceptance as a primary clinical diagnostic tool for EV evaluation. To overcome existing diagnostic challenges, we present the Multi-Organ-cOhesion-Network (MOON), a novel framework enhancing the analysis of critical organ features in NC-CT scans for effective assessment of EV. Drawing inspiration from the thorough assessment practices of radiologists, MOON establishes a cohesive multiorgan analysis model that unifies the imaging features of the related organs of EV, namely esophagus, liver, and spleen. This integration significantly increases the diagnostic accuracy for EV. We have compiled an extensive NC-CT dataset of 1,255 patients diagnosed with EV, spanning three grades of severity. Each case is corroborated by endoscopic diagnostic results. The efficacy of MOON has been substantiated through a validation process involving multi-fold cross-validation on 1,010 cases and an independent test on 245 cases, exhibiting superior diagnostic performance compared to methods focusing solely on the esophagus (for classifying severe grade: AUC of 0.864 versus 0.803, and for moderate to severe grades: AUC of 0.832 versus 0.793). To our knowledge, MOON is the first work to incorporate a synchronized multi-organ NC-CT analysis for EV assessment, providing a more acceptable and minimally invasive alternative for patients compared to traditional endoscopy.
LIDIA: Precise Liver Tumor Diagnosis on Multi-Phase Contrast-Enhanced CT via Iterative Fusion and Asymmetric Contrastive Learning
Jul 18, 2024



Abstract:The early detection and precise diagnosis of liver tumors are tasks of critical clinical value, yet they pose significant challenges due to the high heterogeneity and variability of liver tumors. In this work, a precise LIver tumor DIAgnosis network on multi-phase contrast-enhance CT, named LIDIA, is proposed for real-world scenario. To fully utilize all available phases in contrast-enhanced CT, LIDIA first employs the iterative fusion module to aggregate variable numbers of image phases, thereby capturing the features of lesions at different phases for better tumor diagnosis. To effectively mitigate the high heterogeneity problem of liver tumors, LIDIA incorporates asymmetric contrastive learning to enhance the discriminability between different classes. To evaluate our method, we constructed a large-scale dataset comprising 1,921 patients and 8,138 lesions. LIDIA has achieved an average AUC of 93.6% across eight different types of lesions, demonstrating its effectiveness. Besides, LIDIA also demonstrated strong generalizability with an average AUC of 89.3% when tested on an external cohort of 828 patients.
Boosting Medical Image-based Cancer Detection via Text-guided Supervision from Reports
May 23, 2024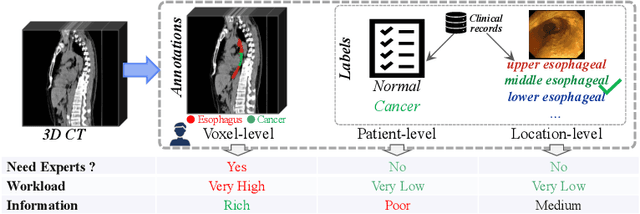
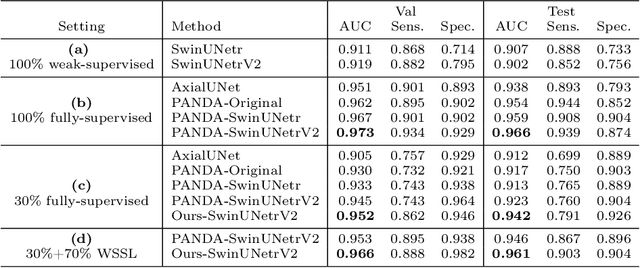
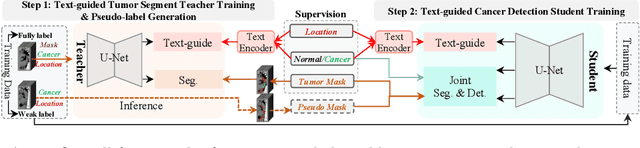
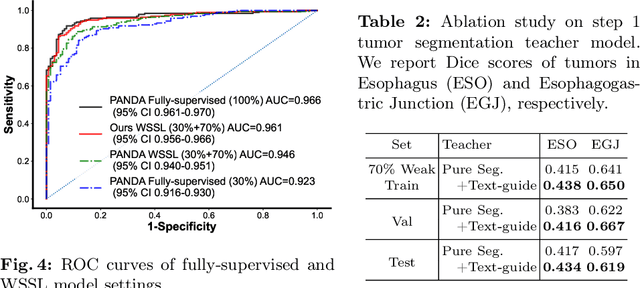
Abstract:The absence of adequately sufficient expert-level tumor annotations hinders the effectiveness of supervised learning based opportunistic cancer screening on medical imaging. Clinical reports (that are rich in descriptive textual details) can offer a "free lunch'' supervision information and provide tumor location as a type of weak label to cope with screening tasks, thus saving human labeling workloads, if properly leveraged. However, predicting cancer only using such weak labels can be very changeling since tumors are usually presented in small anatomical regions compared to the whole 3D medical scans. Weakly semi-supervised learning (WSSL) utilizes a limited set of voxel-level tumor annotations and incorporates alongside a substantial number of medical images that have only off-the-shelf clinical reports, which may strike a good balance between minimizing expert annotation workload and optimizing screening efficacy. In this paper, we propose a novel text-guided learning method to achieve highly accurate cancer detection results. Through integrating diagnostic and tumor location text prompts into the text encoder of a vision-language model (VLM), optimization of weakly supervised learning can be effectively performed in the latent space of VLM, thereby enhancing the stability of training. Our approach can leverage clinical knowledge by large-scale pre-trained VLM to enhance generalization ability, and produce reliable pseudo tumor masks to improve cancer detection. Our extensive quantitative experimental results on a large-scale cancer dataset, including 1,651 unique patients, validate that our approach can reduce human annotation efforts by at least 70% while maintaining comparable cancer detection accuracy to competing fully supervised methods (AUC value 0.961 versus 0.966).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge