Yuxing Tang
Anatomy-Aware Low-Dose CT Denoising via Pretrained Vision Models and Semantic-Guided Contrastive Learning
Aug 11, 2025Abstract:To reduce radiation exposure and improve the diagnostic efficacy of low-dose computed tomography (LDCT), numerous deep learning-based denoising methods have been developed to mitigate noise and artifacts. However, most of these approaches ignore the anatomical semantics of human tissues, which may potentially result in suboptimal denoising outcomes. To address this problem, we propose ALDEN, an anatomy-aware LDCT denoising method that integrates semantic features of pretrained vision models (PVMs) with adversarial and contrastive learning. Specifically, we introduce an anatomy-aware discriminator that dynamically fuses hierarchical semantic features from reference normal-dose CT (NDCT) via cross-attention mechanisms, enabling tissue-specific realism evaluation in the discriminator. In addition, we propose a semantic-guided contrastive learning module that enforces anatomical consistency by contrasting PVM-derived features from LDCT, denoised CT and NDCT, preserving tissue-specific patterns through positive pairs and suppressing artifacts via dual negative pairs. Extensive experiments conducted on two LDCT denoising datasets reveal that ALDEN achieves the state-of-the-art performance, offering superior anatomy preservation and substantially reducing over-smoothing issue of previous work. Further validation on a downstream multi-organ segmentation task (encompassing 117 anatomical structures) affirms the model's ability to maintain anatomical awareness.
Mamba-3D as Masked Autoencoders for Accurate and Data-Efficient Analysis of Medical Ultrasound Videos
Mar 26, 2025Abstract:Ultrasound videos are an important form of clinical imaging data, and deep learning-based automated analysis can improve diagnostic accuracy and clinical efficiency. However, the scarcity of labeled data and the inherent challenges of video analysis have impeded the advancement of related methods. In this work, we introduce E-ViM$^3$, a data-efficient Vision Mamba network that preserves the 3D structure of video data, enhancing long-range dependencies and inductive biases to better model space-time correlations. With our design of Enclosure Global Tokens (EGT), the model captures and aggregates global features more effectively than competing methods. To further improve data efficiency, we employ masked video modeling for self-supervised pre-training, with the proposed Spatial-Temporal Chained (STC) masking strategy designed to adapt to various video scenarios. Experiments demonstrate that E-ViM$^3$ performs as the state-of-the-art in two high-level semantic analysis tasks across four datasets of varying sizes: EchoNet-Dynamic, CAMUS, MICCAI-BUV, and WHBUS. Furthermore, our model achieves competitive performance with limited labels, highlighting its potential impact on real-world clinical applications.
Bootstrapping Chest CT Image Understanding by Distilling Knowledge from X-ray Expert Models
Apr 07, 2024



Abstract:Radiologists highly desire fully automated versatile AI for medical imaging interpretation. However, the lack of extensively annotated large-scale multi-disease datasets has hindered the achievement of this goal. In this paper, we explore the feasibility of leveraging language as a naturally high-quality supervision for chest CT imaging. In light of the limited availability of image-report pairs, we bootstrap the understanding of 3D chest CT images by distilling chest-related diagnostic knowledge from an extensively pre-trained 2D X-ray expert model. Specifically, we propose a language-guided retrieval method to match each 3D CT image with its semantically closest 2D X-ray image, and perform pair-wise and semantic relation knowledge distillation. Subsequently, we use contrastive learning to align images and reports within the same patient while distinguishing them from the other patients. However, the challenge arises when patients have similar semantic diagnoses, such as healthy patients, potentially confusing if treated as negatives. We introduce a robust contrastive learning that identifies and corrects these false negatives. We train our model with over 12,000 pairs of chest CT images and radiology reports. Extensive experiments across multiple scenarios, including zero-shot learning, report generation, and fine-tuning processes, demonstrate the model's feasibility in interpreting chest CT images.
CycleINR: Cycle Implicit Neural Representation for Arbitrary-Scale Volumetric Super-Resolution of Medical Data
Apr 07, 2024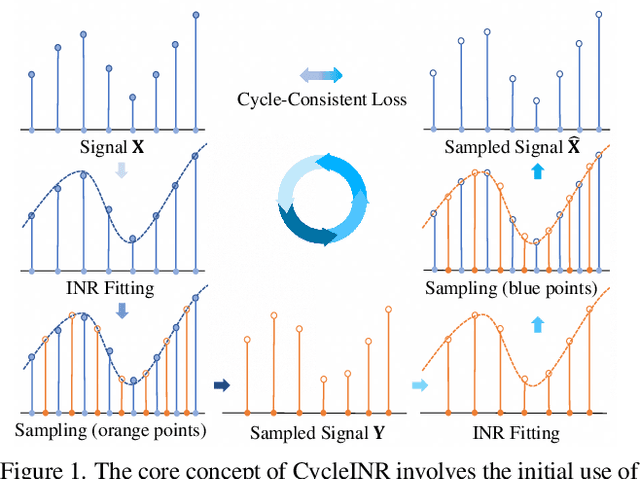
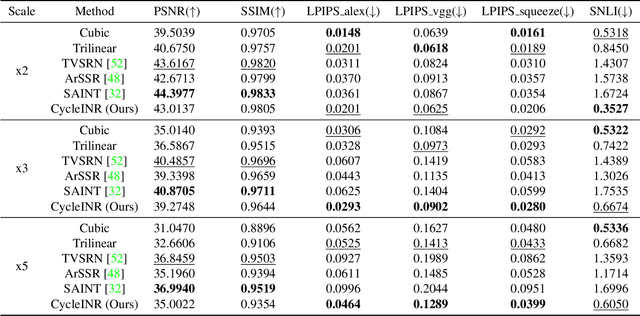
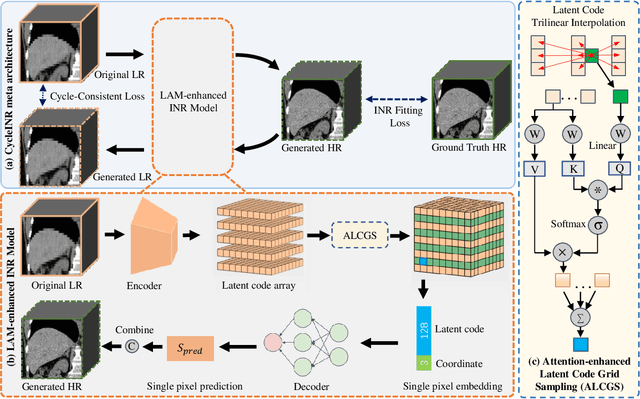
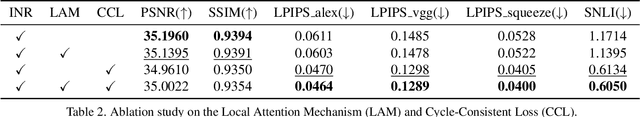
Abstract:In the realm of medical 3D data, such as CT and MRI images, prevalent anisotropic resolution is characterized by high intra-slice but diminished inter-slice resolution. The lowered resolution between adjacent slices poses challenges, hindering optimal viewing experiences and impeding the development of robust downstream analysis algorithms. Various volumetric super-resolution algorithms aim to surmount these challenges, enhancing inter-slice resolution and overall 3D medical imaging quality. However, existing approaches confront inherent challenges: 1) often tailored to specific upsampling factors, lacking flexibility for diverse clinical scenarios; 2) newly generated slices frequently suffer from over-smoothing, degrading fine details, and leading to inter-slice inconsistency. In response, this study presents CycleINR, a novel enhanced Implicit Neural Representation model for 3D medical data volumetric super-resolution. Leveraging the continuity of the learned implicit function, the CycleINR model can achieve results with arbitrary up-sampling rates, eliminating the need for separate training. Additionally, we enhance the grid sampling in CycleINR with a local attention mechanism and mitigate over-smoothing by integrating cycle-consistent loss. We introduce a new metric, Slice-wise Noise Level Inconsistency (SNLI), to quantitatively assess inter-slice noise level inconsistency. The effectiveness of our approach is demonstrated through image quality evaluations on an in-house dataset and a downstream task analysis on the Medical Segmentation Decathlon liver tumor dataset.
Improved Prognostic Prediction of Pancreatic Cancer Using Multi-Phase CT by Integrating Neural Distance and Texture-Aware Transformer
Aug 01, 2023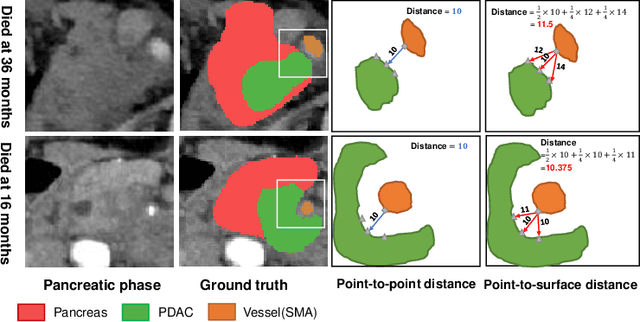
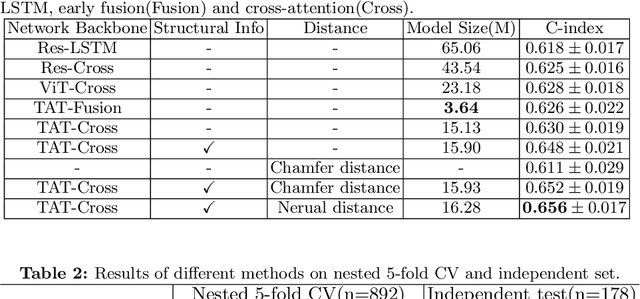
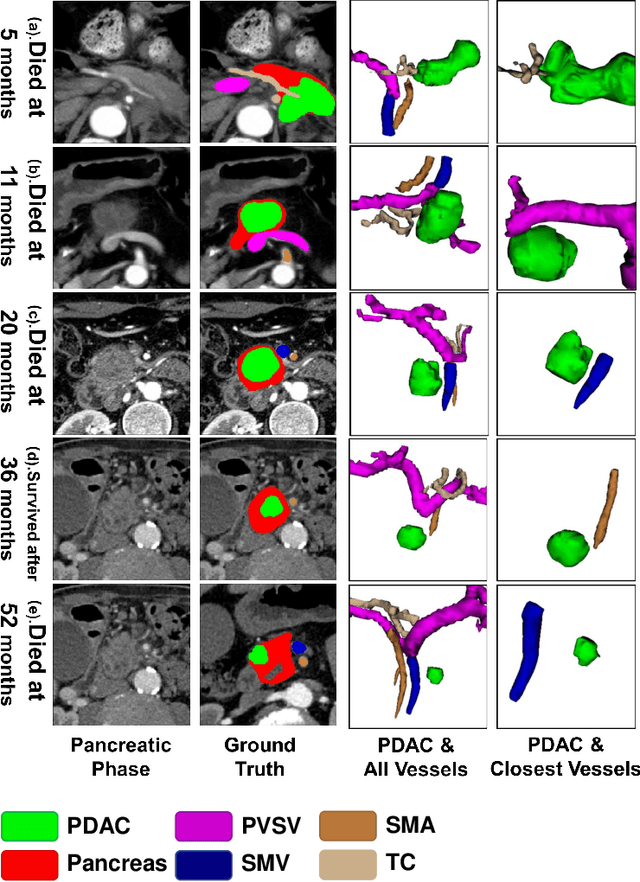
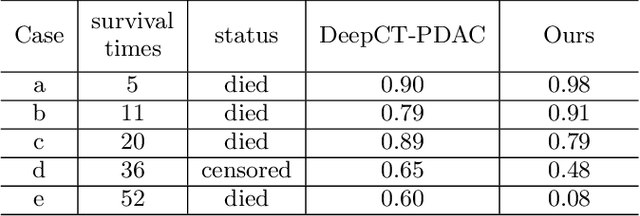
Abstract:Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal cancer in which the tumor-vascular involvement greatly affects the resectability and, thus, overall survival of patients. However, current prognostic prediction methods fail to explicitly and accurately investigate relationships between the tumor and nearby important vessels. This paper proposes a novel learnable neural distance that describes the precise relationship between the tumor and vessels in CT images of different patients, adopting it as a major feature for prognosis prediction. Besides, different from existing models that used CNNs or LSTMs to exploit tumor enhancement patterns on dynamic contrast-enhanced CT imaging, we improved the extraction of dynamic tumor-related texture features in multi-phase contrast-enhanced CT by fusing local and global features using CNN and transformer modules, further enhancing the features extracted across multi-phase CT images. We extensively evaluated and compared the proposed method with existing methods in the multi-center (n=4) dataset with 1,070 patients with PDAC, and statistical analysis confirmed its clinical effectiveness in the external test set consisting of three centers. The developed risk marker was the strongest predictor of overall survival among preoperative factors and it has the potential to be combined with established clinical factors to select patients at higher risk who might benefit from neoadjuvant therapy.
Parse and Recall: Towards Accurate Lung Nodule Malignancy Prediction like Radiologists
Jul 20, 2023



Abstract:Lung cancer is a leading cause of death worldwide and early screening is critical for improving survival outcomes. In clinical practice, the contextual structure of nodules and the accumulated experience of radiologists are the two core elements related to the accuracy of identification of benign and malignant nodules. Contextual information provides comprehensive information about nodules such as location, shape, and peripheral vessels, and experienced radiologists can search for clues from previous cases as a reference to enrich the basis of decision-making. In this paper, we propose a radiologist-inspired method to simulate the diagnostic process of radiologists, which is composed of context parsing and prototype recalling modules. The context parsing module first segments the context structure of nodules and then aggregates contextual information for a more comprehensive understanding of the nodule. The prototype recalling module utilizes prototype-based learning to condense previously learned cases as prototypes for comparative analysis, which is updated online in a momentum way during training. Building on the two modules, our method leverages both the intrinsic characteristics of the nodules and the external knowledge accumulated from other nodules to achieve a sound diagnosis. To meet the needs of both low-dose and noncontrast screening, we collect a large-scale dataset of 12,852 and 4,029 nodules from low-dose and noncontrast CTs respectively, each with pathology- or follow-up-confirmed labels. Experiments on several datasets demonstrate that our method achieves advanced screening performance on both low-dose and noncontrast scenarios.
Towards a Single Unified Model for Effective Detection, Segmentation, and Diagnosis of Eight Major Cancers Using a Large Collection of CT Scans
Jan 28, 2023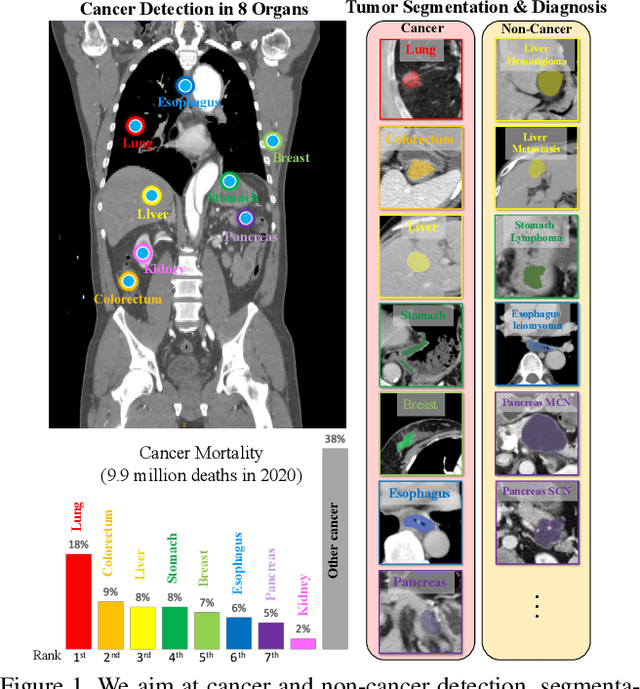
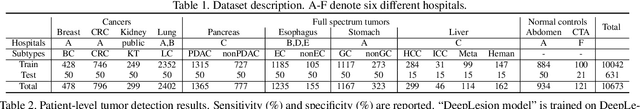
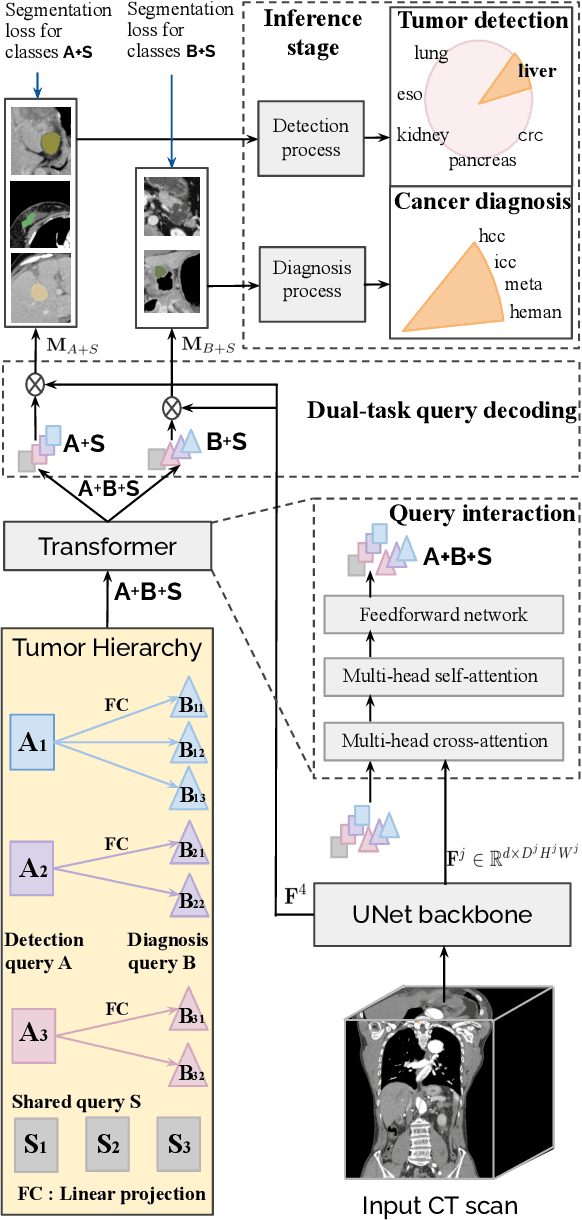
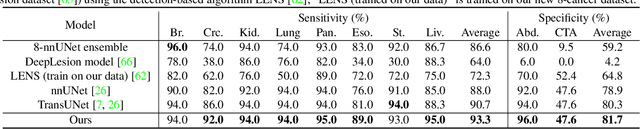
Abstract:Human readers or radiologists routinely perform full-body multi-organ multi-disease detection and diagnosis in clinical practice, while most medical AI systems are built to focus on single organs with a narrow list of a few diseases. This might severely limit AI's clinical adoption. A certain number of AI models need to be assembled non-trivially to match the diagnostic process of a human reading a CT scan. In this paper, we construct a Unified Tumor Transformer (UniT) model to detect (tumor existence and location) and diagnose (tumor characteristics) eight major cancer-prevalent organs in CT scans. UniT is a query-based Mask Transformer model with the output of multi-organ and multi-tumor semantic segmentation. We decouple the object queries into organ queries, detection queries and diagnosis queries, and further establish hierarchical relationships among the three groups. This clinically-inspired architecture effectively assists inter- and intra-organ representation learning of tumors and facilitates the resolution of these complex, anatomically related multi-organ cancer image reading tasks. UniT is trained end-to-end using a curated large-scale CT images of 10,042 patients including eight major types of cancers and occurring non-cancer tumors (all are pathology-confirmed with 3D tumor masks annotated by radiologists). On the test set of 631 patients, UniT has demonstrated strong performance under a set of clinically relevant evaluation metrics, substantially outperforming both multi-organ segmentation methods and an assembly of eight single-organ expert models in tumor detection, segmentation, and diagnosis. Such a unified multi-cancer image reading model (UniT) can significantly reduce the number of false positives produced by combined multi-system models. This moves one step closer towards a universal high-performance cancer screening tool.
E$^2$Net: An Edge Enhanced Network for Accurate Liver and Tumor Segmentation on CT Scans
Jul 19, 2020



Abstract:Developing an effective liver and liver tumor segmentation model from CT scans is very important for the success of liver cancer diagnosis, surgical planning and cancer treatment. In this work, we propose a two-stage framework for 2D liver and tumor segmentation. The first stage is a coarse liver segmentation network, while the second stage is an edge enhanced network (E$^2$Net) for more accurate liver and tumor segmentation. E$^2$Net explicitly models complementary objects (liver and tumor) and their edge information within the network to preserve the organ and lesion boundaries. We introduce an edge prediction module in E$^2$Net and design an edge distance map between liver and tumor boundaries, which is used as an extra supervision signal to train the edge enhanced network. We also propose a deep cross feature fusion module to refine multi-scale features from both objects and their edges. E$^2$Net is more easily and efficiently trained with a small labeled dataset, and it can be trained/tested on the original 2D CT slices (resolve resampling error issue in 3D models). The proposed framework has shown superior performance on both liver and liver tumor segmentation compared to several state-of-the-art 2D, 3D and 2D/3D hybrid frameworks.
Cross-Domain Medical Image Translation by Shared Latent Gaussian Mixture Model
Jul 14, 2020



Abstract:Current deep learning based segmentation models often generalize poorly between domains due to insufficient training data. In real-world clinical applications, cross-domain image analysis tools are in high demand since medical images from different domains are often needed to achieve a precise diagnosis. An important example in radiology is generalizing from non-contrast CT to contrast enhanced CTs. Contrast enhanced CT scans at different phases are used to enhance certain pathologies or organs. Many existing cross-domain image-to-image translation models have been shown to improve cross-domain segmentation of large organs. However, such models lack the ability to preserve fine structures during the translation process, which is significant for many clinical applications, such as segmenting small calcified plaques in the aorta and pelvic arteries. In order to preserve fine structures during medical image translation, we propose a patch-based model using shared latent variables from a Gaussian mixture model. We compare our image translation framework to several state-of-the-art methods on cross-domain image translation and show our model does a better job preserving fine structures. The superior performance of our model is verified by performing two tasks with the translated images - detection and segmentation of aortic plaques and pancreas segmentation. We expect the utility of our framework will extend to other problems beyond segmentation due to the improved quality of the generated images and enhanced ability to preserve small structures.
Bone Suppression on Chest Radiographs With Adversarial Learning
Feb 08, 2020



Abstract:Dual-energy (DE) chest radiography provides the capability of selectively imaging two clinically relevant materials, namely soft tissues, and osseous structures, to better characterize a wide variety of thoracic pathology and potentially improve diagnosis in posteroanterior (PA) chest radiographs. However, DE imaging requires specialized hardware and a higher radiation dose than conventional radiography, and motion artifacts sometimes happen due to involuntary patient motion. In this work, we learn the mapping between conventional radiographs and bone suppressed radiographs. Specifically, we propose to utilize two variations of generative adversarial networks (GANs) for image-to-image translation between conventional and bone suppressed radiographs obtained by DE imaging technique. We compare the effectiveness of training with patient-wisely paired and unpaired radiographs. Experiments show both training strategies yield "radio-realistic'' radiographs with suppressed bony structures and few motion artifacts on a hold-out test set. While training with paired images yields slightly better performance than that of unpaired images when measuring with two objective image quality metrics, namely Structural Similarity Index (SSIM) and Peak Signal-to-Noise Ratio (PSNR), training with unpaired images demonstrates better generalization ability on unseen anteroposterior (AP) radiographs than paired training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge