Daniel C. Elton
Lymph Node Detection in T2 MRI with Transformers
Nov 09, 2021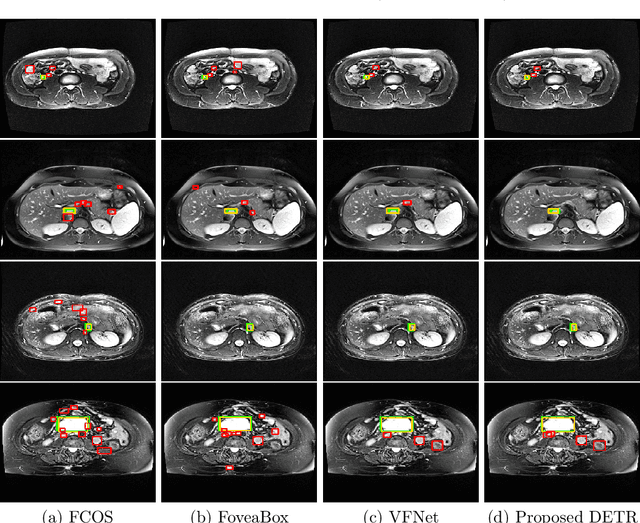
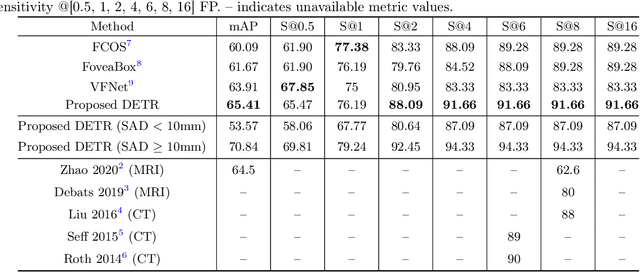
Abstract:Identification of lymph nodes (LN) in T2 Magnetic Resonance Imaging (MRI) is an important step performed by radiologists during the assessment of lymphoproliferative diseases. The size of the nodes play a crucial role in their staging, and radiologists sometimes use an additional contrast sequence such as diffusion weighted imaging (DWI) for confirmation. However, lymph nodes have diverse appearances in T2 MRI scans, making it tough to stage for metastasis. Furthermore, radiologists often miss smaller metastatic lymph nodes over the course of a busy day. To deal with these issues, we propose to use the DEtection TRansformer (DETR) network to localize suspicious metastatic lymph nodes for staging in challenging T2 MRI scans acquired by different scanners and exam protocols. False positives (FP) were reduced through a bounding box fusion technique, and a precision of 65.41\% and sensitivity of 91.66\% at 4 FP per image was achieved. To the best of our knowledge, our results improve upon the current state-of-the-art for lymph node detection in T2 MRI scans.
Induction, Popper, and machine learning
Oct 02, 2021
Abstract:Francis Bacon popularized the idea that science is based on a process of induction by which repeated observations are, in some unspecified way, generalized to theories based on the assumption that the future resembles the past. This idea was criticized by Hume and others as untenable leading to the famous problem of induction. It wasn't until the work of Karl Popper that this problem was solved, by demonstrating that induction is not the basis for science and that the development of scientific knowledge is instead based on the same principles as biological evolution. Today, machine learning is also taught as being rooted in induction from big data. Solomonoff induction implemented in an idealized Bayesian agent (Hutter's AIXI) is widely discussed and touted as a framework for understanding AI algorithms, even though real-world attempts to implement something like AIXI immediately encounter fatal problems. In this paper, we contrast frameworks based on induction with Donald T. Campbell's universal Darwinism. We show that most AI algorithms in use today can be understood as using an evolutionary trial and error process searching over a solution space. In this work we argue that a universal Darwinian framework provides a better foundation for understanding AI systems. Moreover, at a more meta level the process of development of all AI algorithms can be understood under the framework of universal Darwinism.
Applying Deutsch's concept of good explanations to artificial intelligence and neuroscience -- an initial exploration
Dec 24, 2020Abstract:Artificial intelligence has made great strides since the deep learning revolution, but AI systems still struggle to extrapolate outside of their training data and adapt to new situations. For inspiration we look to the domain of science, where scientists have been able to develop theories which show remarkable ability to extrapolate and sometimes predict the existence of phenomena which have never been observed before. According to David Deutsch, this type of extrapolation, which he calls "reach", is due to scientific theories being hard to vary. In this work we investigate Deutsch's hard-to-vary principle and how it relates to more formalized principles in deep learning such as the bias-variance trade-off and Occam's razor. We distinguish internal variability, how much a model/theory can be varied internally while still yielding the same predictions, with external variability, which is how much a model must be varied to accurately predict new, out-of-distribution data. We discuss how to measure internal variability using the size of the Rashomon set and how to measure external variability using Kolmogorov complexity. We explore what role hard-to-vary explanations play in intelligence by looking at the human brain and distinguish two learning systems in the brain. The first system operates similar to deep learning and likely underlies most of perception and motor control while the second is a more creative system capable of generating hard-to-vary explanations of the world. We argue that figuring out how replicate this second system, which is capable of generating hard-to-vary explanations, is a key challenge which needs to be solved in order to realize artificial general intelligence. We make contact with the framework of Popperian epistemology which rejects induction and asserts that knowledge generation is an evolutionary process which proceeds through conjecture and refutation.
Deep Small Bowel Segmentation with Cylindrical Topological Constraints
Jul 16, 2020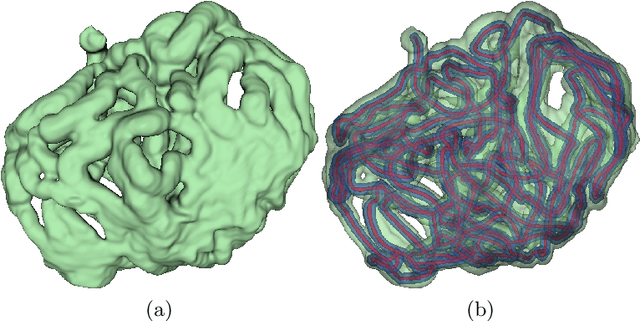
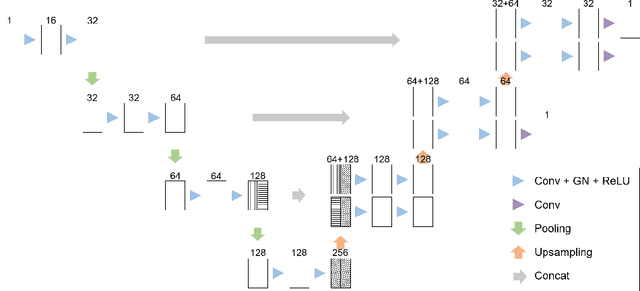

Abstract:We present a novel method for small bowel segmentation where a cylindrical topological constraint based on persistent homology is applied. To address the touching issue which could break the applied constraint, we propose to augment a network with an additional branch to predict an inner cylinder of the small bowel. Since the inner cylinder is free of the touching issue, a cylindrical shape constraint applied on this augmented branch guides the network to generate a topologically correct segmentation. For strict evaluation, we achieved an abdominal computed tomography dataset with dense segmentation ground-truths. The proposed method showed clear improvements in terms of four different metrics compared to the baseline method, and also showed the statistical significance from a paired t-test.
Cross-Domain Medical Image Translation by Shared Latent Gaussian Mixture Model
Jul 14, 2020



Abstract:Current deep learning based segmentation models often generalize poorly between domains due to insufficient training data. In real-world clinical applications, cross-domain image analysis tools are in high demand since medical images from different domains are often needed to achieve a precise diagnosis. An important example in radiology is generalizing from non-contrast CT to contrast enhanced CTs. Contrast enhanced CT scans at different phases are used to enhance certain pathologies or organs. Many existing cross-domain image-to-image translation models have been shown to improve cross-domain segmentation of large organs. However, such models lack the ability to preserve fine structures during the translation process, which is significant for many clinical applications, such as segmenting small calcified plaques in the aorta and pelvic arteries. In order to preserve fine structures during medical image translation, we propose a patch-based model using shared latent variables from a Gaussian mixture model. We compare our image translation framework to several state-of-the-art methods on cross-domain image translation and show our model does a better job preserving fine structures. The superior performance of our model is verified by performing two tasks with the translated images - detection and segmentation of aortic plaques and pancreas segmentation. We expect the utility of our framework will extend to other problems beyond segmentation due to the improved quality of the generated images and enhanced ability to preserve small structures.
Image Translation by Latent Union of Subspaces for Cross-Domain Plaque Detection
May 22, 2020

Abstract:Calcified plaque in the aorta and pelvic arteries is associated with coronary artery calcification and is a strong predictor of heart attack. Current calcified plaque detection models show poor generalizability to different domains (ie. pre-contrast vs. post-contrast CT scans). Many recent works have shown how cross domain object detection can be improved using an image translation model which translates between domains using a single shared latent space. However, while current image translation models do a good job preserving global/intermediate level structures they often have trouble preserving tiny structures. In medical imaging applications, preserving small structures is important since these structures can carry information which is highly relevant for disease diagnosis. Recent works on image reconstruction show that complex real-world images are better reconstructed using a union of subspaces approach. Since small image patches are used to train the image translation model, it makes sense to enforce that each patch be represented by a linear combination of subspaces which may correspond to the different parts of the body present in that patch. Motivated by this, we propose an image translation network using a shared union of subspaces constraint and show our approach preserves subtle structures (plaques) better than the conventional method. We further applied our method to a cross domain plaque detection task and show significant improvement compared to the state-of-the art method.
Self-explaining AI as an alternative to interpretable AI
Feb 29, 2020
Abstract:The ability to explain decisions made by AI systems is highly sought after, especially in domains where human lives are at stake such as medicine or autonomous vehicles. While it is always possible to approximate the input-output relations of deep neural networks with human-understandable rules or a post-hoc model, the discovery of the double descent phenomena suggests that no such approximation will ever map onto the actual mechanistic functioning of deep neural networks. Double descent indicates that deep neural networks typically operate by smoothly interpolating between data points rather than by extracting a few high level rules. As a result neural networks trained on complex real world data are inherently hard to interpret and prone to failure if used outside their domain of applicability (ie, for extrapolation). To show how we might be able to trust AI despite these problems, we introduce the concept of self-explaining AI. Self-explaining AIs are capable of providing a human-understandable explanation of each decision along with confidence levels for both the decision and explanation. Some difficulties to this approach along with possible solutions are sketched. Finally, we argue it is also important that AI systems warn their user when they are asked to perform outside their domain of applicability.
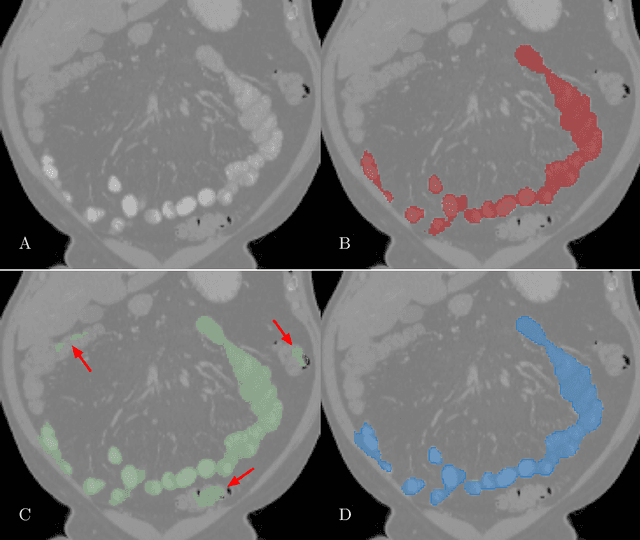
Accurately identifying vertebral levels in large datasets
Jan 28, 2020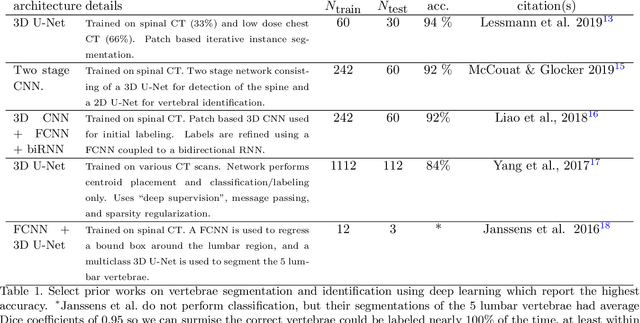
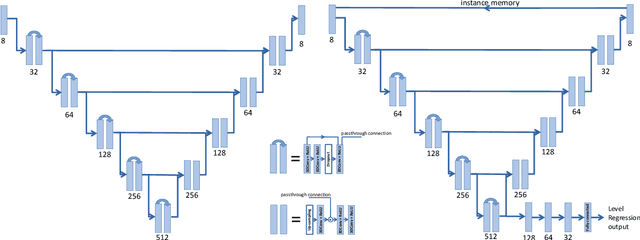
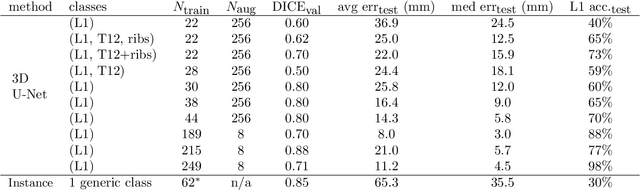
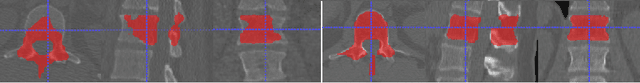
Abstract:The vertebral levels of the spine provide a useful coordinate system when making measurements of plaque, muscle, fat, and bone mineral density. Correctly classifying vertebral levels with high accuracy is challenging due to the similar appearance of each vertebra, the curvature of the spine, and the possibility of anomalies such as fractured vertebrae, implants, lumbarization of the sacrum, and sacralization of L5. The goal of this work is to develop a system that can accurately and robustly identify the L1 level in large heterogeneous datasets. The first approach we study is using a 3D U-Net to segment the L1 vertebra directly using the entire scan volume to provide context. We also tested models for two class segmentation of L1 and T12 and a three class segmentation of L1, T12 and the rib attached to T12. By increasing the number of training examples to 249 scans using pseudo-segmentations from an in-house segmentation tool we were able to achieve 98% accuracy with respect to identifying the L1 vertebra, with an average error of 4.5 mm in the craniocaudal level. We next developed an algorithm which performs iterative instance segmentation and classification of the entire spine with a 3D U-Net. We found the instance based approach was able to yield better segmentations of nearly the entire spine, but had lower classification accuracy for L1.
Deep learning for molecular generation and optimization - a review of the state of the art
Mar 11, 2019
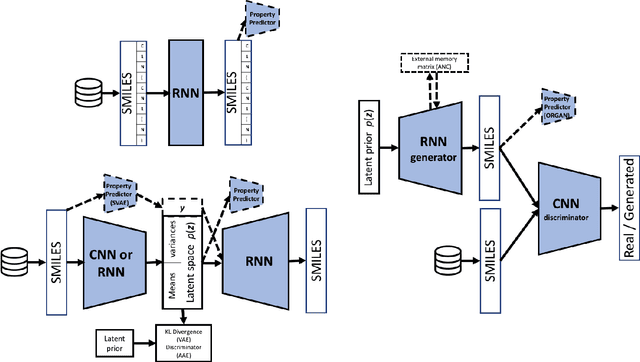
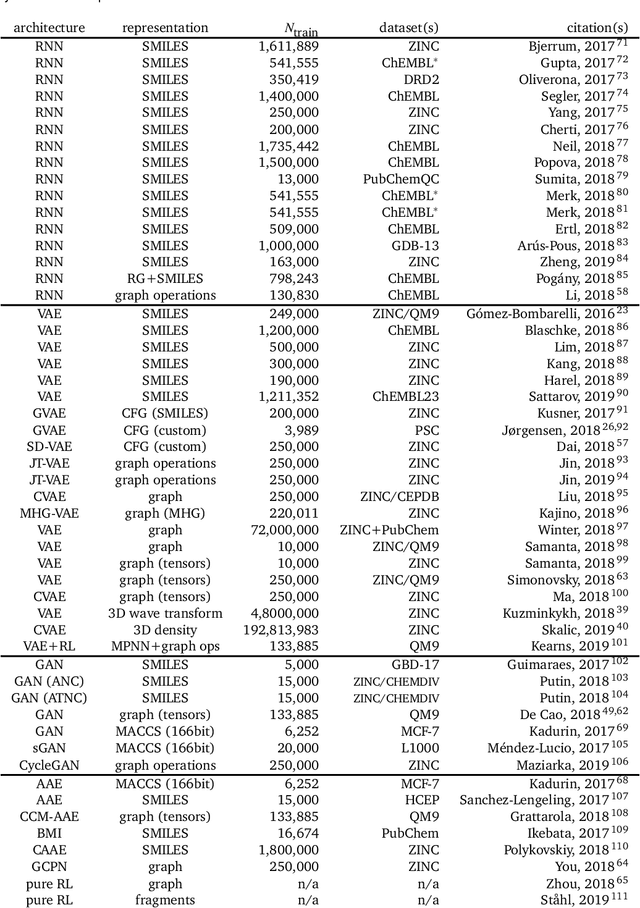
Abstract:In the space of only a few years, deep generative modeling has revolutionized how we think of artificial creativity, yielding autonomous systems which produce original images, music, and text. Inspired by these successes, researchers are now applying deep generative modeling techniques to the generation and optimization of molecules - in our review we found 45 papers on the subject published in the past two years. These works point to a future where such systems will be used to generate lead molecules, greatly reducing resources spent downstream synthesizing and characterizing bad leads in the lab. In this review we survey the increasingly complex landscape of models and representation schemes that have been proposed. The four classes of techniques we describe are recursive neural networks, autoencoders, generative adversarial networks, and reinforcement learning. After first discussing some of the mathematical fundamentals of each technique, we draw high level connections and comparisons with other techniques and expose the pros and cons of each. Several important high level themes emerge as a result of this work, including the shift away from the SMILES string representation of molecules towards more sophisticated representations such as graph grammars and 3D representations, the importance of reward function design, the need for better standards for benchmarking and testing, and the benefits of adversarial training and reinforcement learning over maximum likelihood based training.
Using natural language processing techniques to extract information on the properties and functionalities of energetic materials from large text corpora
Mar 01, 2019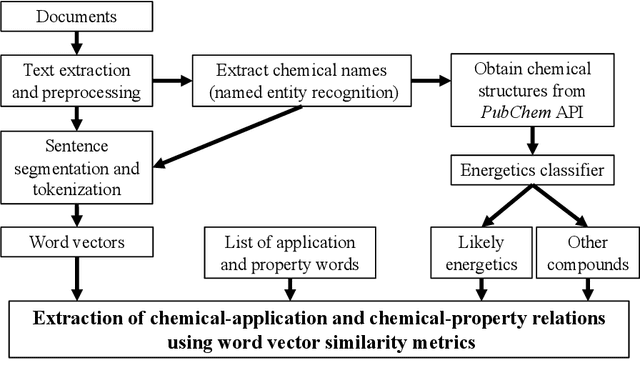
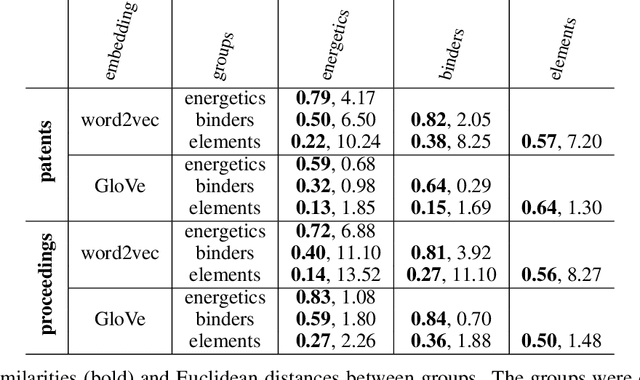
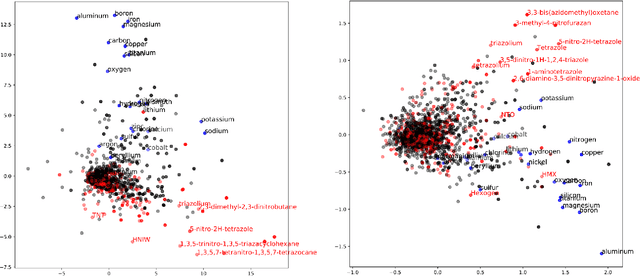

Abstract:The number of scientific journal articles and reports being published about energetic materials every year is growing exponentially, and therefore extracting relevant information and actionable insights from the latest research is becoming a considerable challenge. In this work we explore how techniques from natural language processing and machine learning can be used to automatically extract chemical insights from large collections of documents. We first describe how to download and process documents from a variety of sources - journal articles, conference proceedings (including NTREM), the US Patent & Trademark Office, and the Defense Technical Information Center archive on archive.org. We present a custom NLP pipeline which uses open source NLP tools to identify the names of chemical compounds and relates them to function words ("underwater", "rocket", "pyrotechnic") and property words ("elastomer", "non-toxic"). After explaining how word embeddings work we compare the utility of two popular word embeddings - word2vec and GloVe. Chemical-chemical and chemical-application relationships are obtained by doing computations with word vectors. We show that word embeddings capture latent information about energetic materials, so that related materials appear close together in the word embedding space.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge