Yifan Peng
Ret.
A General Model for Retinal Segmentation and Quantification
Jan 31, 2026Abstract:Retinal imaging is fast, non-invasive, and widely available, offering quantifiable structural and vascular signals for ophthalmic and systemic health assessment. This accessibility creates an opportunity to study how quantitative retinal phenotypes relate to ocular and systemic diseases. However, such analyses remain difficult at scale due to the limited availability of public multi-label datasets and the lack of a unified segmentation-to-quantification pipeline. We present RetSAM, a general retinal segmentation and quantification framework for fundus imaging. It delivers robust multi-target segmentation and standardized biomarker extraction, supporting downstream ophthalmologic studies and oculomics correlation analyses. Trained on over 200,000 fundus images, RetSAM supports three task categories and segments five anatomical structures, four retinal phenotypic patterns, and more than 20 distinct lesion types. It converts these segmentation results into over 30 standardized biomarkers that capture structural morphology, vascular geometry, and degenerative changes. Trained with a multi-stage strategy using both private and public fundus data, RetSAM achieves superior segmentation performance on 17 public datasets. It improves on prior best methods by 3.9 percentage points in DSC on average, with up to 15 percentage points on challenging multi-task benchmarks, and generalizes well across diverse populations, imaging devices, and clinical settings. The resulting biomarkers enable systematic correlation analyses across major ophthalmic diseases, including diabetic retinopathy, age-related macular degeneration, glaucoma, and pathologic myopia. Together, RetSAM transforms fundus images into standardized, interpretable quantitative phenotypes, enabling large-scale ophthalmic research and translation.
Scaling Medical Reasoning Verification via Tool-Integrated Reinforcement Learning
Jan 28, 2026Abstract:Large language models have achieved strong performance on medical reasoning benchmarks, yet their deployment in clinical settings demands rigorous verification to ensure factual accuracy. While reward models offer a scalable approach for reasoning trace verification, existing methods face two limitations: they produce only scalar reward values without explicit justification, and they rely on single-pass retrieval that precludes adaptive knowledge access as verification unfolds. We introduce $\method$, an agentic framework that addresses these limitations by training medical reasoning verifiers to iteratively query external medical corpora during evaluation. Our approach combines tool-augmented verification with an iterative reinforcement learning paradigm that requires only trace-level supervision, alongside an adaptive curriculum mechanism that dynamically adjusts training data distribution. Across four medical reasoning benchmarks, $\method$ achieves substantial gains over existing methods, improving MedQA accuracy by 23.5% and MedXpertQA by 32.0% relative to the base generator in particular. Crucially, $\method$ demonstrates an $\mathbf{8\times}$ reduction in sampling budget requirement compared to prior reward model baselines. These findings establish that grounding verification in dynamically retrieved evidence offers a principled path toward more reliable medical reasoning systems.
Establishing dermatopathology encyclopedia DermpathNet with Artificial Intelligence-Based Workflow
Jan 27, 2026Abstract:Accessing high-quality, open-access dermatopathology image datasets for learning and cross-referencing is a common challenge for clinicians and dermatopathology trainees. To establish a comprehensive open-access dermatopathology dataset for educational, cross-referencing, and machine-learning purposes, we employed a hybrid workflow to curate and categorize images from the PubMed Central (PMC) repository. We used specific keywords to extract relevant images, and classified them using a novel hybrid method that combined deep learning-based image modality classification with figure caption analyses. Validation on 651 manually annotated images demonstrated the robustness of our workflow, with an F-score of 89.6\% for the deep learning approach, 61.0\% for the keyword-based retrieval method, and 90.4\% for the hybrid approach. We retrieved over 7,772 images across 166 diagnoses and released this fully annotated dataset, reviewed by board-certified dermatopathologists. Using our dataset as a challenging task, we found the current image analysis algorithm from OpenAI inadequate for analyzing dermatopathology images. In conclusion, we have developed a large, peer-reviewed, open-access dermatopathology image dataset, DermpathNet, which features a semi-automated curation workflow.
RSNA Large Language Model Benchmark Dataset for Chest Radiographs of Cardiothoracic Disease: Radiologist Evaluation and Validation Enhanced by AI Labels (REVEAL-CXR)
Jan 21, 2026Abstract:Multimodal large language models have demonstrated comparable performance to that of radiology trainees on multiple-choice board-style exams. However, to develop clinically useful multimodal LLM tools, high-quality benchmarks curated by domain experts are essential. To curate released and holdout datasets of 100 chest radiographic studies each and propose an artificial intelligence (AI)-assisted expert labeling procedure to allow radiologists to label studies more efficiently. A total of 13,735 deidentified chest radiographs and their corresponding reports from the MIDRC were used. GPT-4o extracted abnormal findings from the reports, which were then mapped to 12 benchmark labels with a locally hosted LLM (Phi-4-Reasoning). From these studies, 1,000 were sampled on the basis of the AI-suggested benchmark labels for expert review; the sampling algorithm ensured that the selected studies were clinically relevant and captured a range of difficulty levels. Seventeen chest radiologists participated, and they marked "Agree all", "Agree mostly" or "Disagree" to indicate their assessment of the correctness of the LLM suggested labels. Each chest radiograph was evaluated by three experts. Of these, at least two radiologists selected "Agree All" for 381 radiographs. From this set, 200 were selected, prioritizing those with less common or multiple finding labels, and divided into 100 released radiographs and 100 reserved as the holdout dataset. The holdout dataset is used exclusively by RSNA to independently evaluate different models. A benchmark of 200 chest radiographic studies with 12 benchmark labels was created and made publicly available https://imaging.rsna.org, with each chest radiograph verified by three radiologists. In addition, an AI-assisted labeling procedure was developed to help radiologists label at scale, minimize unnecessary omissions, and support a semicollaborative environment.
CPGPrompt: Translating Clinical Guidelines into LLM-Executable Decision Support
Jan 07, 2026Abstract:Clinical practice guidelines (CPGs) provide evidence-based recommendations for patient care; however, integrating them into Artificial Intelligence (AI) remains challenging. Previous approaches, such as rule-based systems, face significant limitations, including poor interpretability, inconsistent adherence to guidelines, and narrow domain applicability. To address this, we develop and validate CPGPrompt, an auto-prompting system that converts narrative clinical guidelines into large language models (LLMs). Our framework translates CPGs into structured decision trees and utilizes an LLM to dynamically navigate them for patient case evaluation. Synthetic vignettes were generated across three domains (headache, lower back pain, and prostate cancer) and distributed into four categories to test different decision scenarios. System performance was assessed on both binary specialty-referral decisions and fine-grained pathway-classification tasks. The binary specialty referral classification achieved consistently strong performance across all domains (F1: 0.85-1.00), with high recall (1.00 $\pm$ 0.00). In contrast, multi-class pathway assignment showed reduced performance, with domain-specific variations: headache (F1: 0.47), lower back pain (F1: 0.72), and prostate cancer (F1: 0.77). Domain-specific performance differences reflected the structure of each guideline. The headache guideline highlighted challenges with negation handling. The lower back pain guideline required temporal reasoning. In contrast, prostate cancer pathways benefited from quantifiable laboratory tests, resulting in more reliable decision-making.
Toward Global Large Language Models in Medicine
Jan 05, 2026Abstract:Despite continuous advances in medical technology, the global distribution of health care resources remains uneven. The development of large language models (LLMs) has transformed the landscape of medicine and holds promise for improving health care quality and expanding access to medical information globally. However, existing LLMs are primarily trained on high-resource languages, limiting their applicability in global medical scenarios. To address this gap, we constructed GlobMed, a large multilingual medical dataset, containing over 500,000 entries spanning 12 languages, including four low-resource languages. Building on this, we established GlobMed-Bench, which systematically assesses 56 state-of-the-art proprietary and open-weight LLMs across multiple multilingual medical tasks, revealing significant performance disparities across languages, particularly for low-resource languages. Additionally, we introduced GlobMed-LLMs, a suite of multilingual medical LLMs trained on GlobMed, with parameters ranging from 1.7B to 8B. GlobMed-LLMs achieved an average performance improvement of over 40% relative to baseline models, with a more than threefold increase in performance on low-resource languages. Together, these resources provide an important foundation for advancing the equitable development and application of LLMs globally, enabling broader language communities to benefit from technological advances.
A Disease-Aware Dual-Stage Framework for Chest X-ray Report Generation
Nov 15, 2025Abstract:Radiology report generation from chest X-rays is an important task in artificial intelligence with the potential to greatly reduce radiologists' workload and shorten patient wait times. Despite recent advances, existing approaches often lack sufficient disease-awareness in visual representations and adequate vision-language alignment to meet the specialized requirements of medical image analysis. As a result, these models usually overlook critical pathological features on chest X-rays and struggle to generate clinically accurate reports. To address these limitations, we propose a novel dual-stage disease-aware framework for chest X-ray report generation. In Stage~1, our model learns Disease-Aware Semantic Tokens (DASTs) corresponding to specific pathology categories through cross-attention mechanisms and multi-label classification, while simultaneously aligning vision and language representations via contrastive learning. In Stage~2, we introduce a Disease-Visual Attention Fusion (DVAF) module to integrate disease-aware representations with visual features, along with a Dual-Modal Similarity Retrieval (DMSR) mechanism that combines visual and disease-specific similarities to retrieve relevant exemplars, providing contextual guidance during report generation. Extensive experiments on benchmark datasets (i.e., CheXpert Plus, IU X-ray, and MIMIC-CXR) demonstrate that our disease-aware framework achieves state-of-the-art performance in chest X-ray report generation, with significant improvements in clinical accuracy and linguistic quality.
A Multi-agent Large Language Model Framework to Automatically Assess Performance of a Clinical AI Triage Tool
Oct 30, 2025Abstract:Purpose: The purpose of this study was to determine if an ensemble of multiple LLM agents could be used collectively to provide a more reliable assessment of a pixel-based AI triage tool than a single LLM. Methods: 29,766 non-contrast CT head exams from fourteen hospitals were processed by a commercial intracranial hemorrhage (ICH) AI detection tool. Radiology reports were analyzed by an ensemble of eight open-source LLM models and a HIPAA compliant internal version of GPT-4o using a single multi-shot prompt that assessed for presence of ICH. 1,726 examples were manually reviewed. Performance characteristics of the eight open-source models and consensus were compared to GPT-4o. Three ideal consensus LLM ensembles were tested for rating the performance of the triage tool. Results: The cohort consisted of 29,766 head CTs exam-report pairs. The highest AUC performance was achieved with llama3.3:70b and GPT-4o (AUC= 0.78). The average precision was highest for Llama3.3:70b and GPT-4o (AP=0.75 & 0.76). Llama3.3:70b had the highest F1 score (0.81) and recall (0.85), greater precision (0.78), specificity (0.72), and MCC (0.57). Using MCC (95% CI) the ideal combination of LLMs were: Full-9 Ensemble 0.571 (0.552-0.591), Top-3 Ensemble 0.558 (0.537-0.579), Consensus 0.556 (0.539-0.574), and GPT4o 0.522 (0.500-0.543). No statistically significant differences were observed between Top-3, Full-9, and Consensus (p > 0.05). Conclusion: An ensemble of medium to large sized open-source LLMs provides a more consistent and reliable method to derive a ground truth retrospective evaluation of a clinical AI triage tool over a single LLM alone.
Two-Stage Decoupling Framework for Variable-Length Glaucoma Prognosis
Sep 15, 2025Abstract:Glaucoma is one of the leading causes of irreversible blindness worldwide. Glaucoma prognosis is essential for identifying at-risk patients and enabling timely intervention to prevent blindness. Many existing approaches rely on historical sequential data but are constrained by fixed-length inputs, limiting their flexibility. Additionally, traditional glaucoma prognosis methods often employ end-to-end models, which struggle with the limited size of glaucoma datasets. To address these challenges, we propose a Two-Stage Decoupling Framework (TSDF) for variable-length glaucoma prognosis. In the first stage, we employ a feature representation module that leverages self-supervised learning to aggregate multiple glaucoma datasets for training, disregarding differences in their supervisory information. This approach enables datasets of varying sizes to learn better feature representations. In the second stage, we introduce a temporal aggregation module that incorporates an attention-based mechanism to process sequential inputs of varying lengths, ensuring flexible and efficient utilization of all available data. This design significantly enhances model performance while maintaining a compact parameter size. Extensive experiments on two benchmark glaucoma datasets:the Ocular Hypertension Treatment Study (OHTS) and the Glaucoma Real-world Appraisal Progression Ensemble (GRAPE),which differ significantly in scale and clinical settings,demonstrate the effectiveness and robustness of our approach.
EventTracer: Fast Path Tracing-based Event Stream Rendering
Aug 25, 2025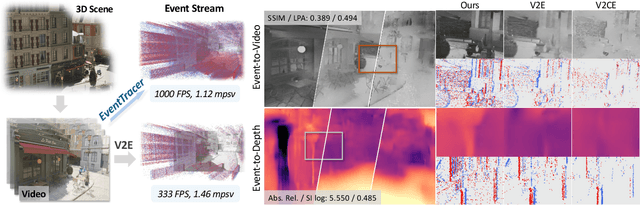

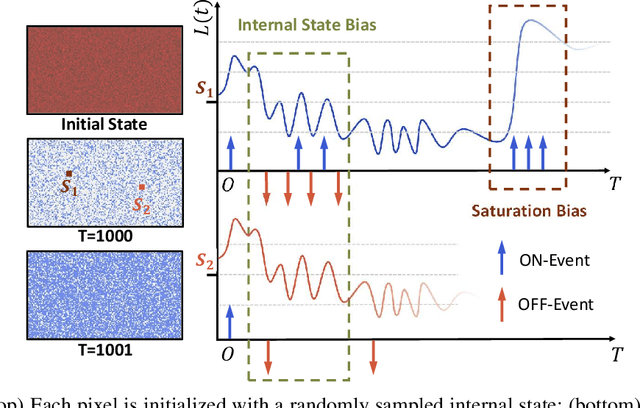
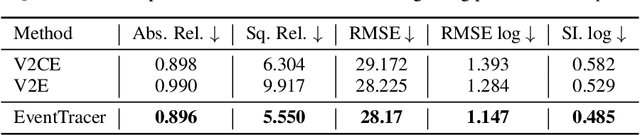
Abstract:Simulating event streams from 3D scenes has become a common practice in event-based vision research, as it meets the demand for large-scale, high temporal frequency data without setting up expensive hardware devices or undertaking extensive data collections. Yet existing methods in this direction typically work with noiseless RGB frames that are costly to render, and therefore they can only achieve a temporal resolution equivalent to 100-300 FPS, far lower than that of real-world event data. In this work, we propose EventTracer, a path tracing-based rendering pipeline that simulates high-fidelity event sequences from complex 3D scenes in an efficient and physics-aware manner. Specifically, we speed up the rendering process via low sample-per-pixel (SPP) path tracing, and train a lightweight event spiking network to denoise the resulting RGB videos into realistic event sequences. To capture the physical properties of event streams, the network is equipped with a bipolar leaky integrate-and-fired (BiLIF) spiking unit and trained with a bidirectional earth mover distance (EMD) loss. Our EventTracer pipeline runs at a speed of about 4 minutes per second of 720p video, and it inherits the merit of accurate spatiotemporal modeling from its path tracing backbone. We show in two downstream tasks that EventTracer captures better scene details and demonstrates a greater similarity to real-world event data than other event simulators, which establishes it as a promising tool for creating large-scale event-RGB datasets at a low cost, narrowing the sim-to-real gap in event-based vision, and boosting various application scenarios such as robotics, autonomous driving, and VRAR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge