Mingquan Lin
A General Model for Retinal Segmentation and Quantification
Jan 31, 2026Abstract:Retinal imaging is fast, non-invasive, and widely available, offering quantifiable structural and vascular signals for ophthalmic and systemic health assessment. This accessibility creates an opportunity to study how quantitative retinal phenotypes relate to ocular and systemic diseases. However, such analyses remain difficult at scale due to the limited availability of public multi-label datasets and the lack of a unified segmentation-to-quantification pipeline. We present RetSAM, a general retinal segmentation and quantification framework for fundus imaging. It delivers robust multi-target segmentation and standardized biomarker extraction, supporting downstream ophthalmologic studies and oculomics correlation analyses. Trained on over 200,000 fundus images, RetSAM supports three task categories and segments five anatomical structures, four retinal phenotypic patterns, and more than 20 distinct lesion types. It converts these segmentation results into over 30 standardized biomarkers that capture structural morphology, vascular geometry, and degenerative changes. Trained with a multi-stage strategy using both private and public fundus data, RetSAM achieves superior segmentation performance on 17 public datasets. It improves on prior best methods by 3.9 percentage points in DSC on average, with up to 15 percentage points on challenging multi-task benchmarks, and generalizes well across diverse populations, imaging devices, and clinical settings. The resulting biomarkers enable systematic correlation analyses across major ophthalmic diseases, including diabetic retinopathy, age-related macular degeneration, glaucoma, and pathologic myopia. Together, RetSAM transforms fundus images into standardized, interpretable quantitative phenotypes, enabling large-scale ophthalmic research and translation.
Establishing dermatopathology encyclopedia DermpathNet with Artificial Intelligence-Based Workflow
Jan 27, 2026Abstract:Accessing high-quality, open-access dermatopathology image datasets for learning and cross-referencing is a common challenge for clinicians and dermatopathology trainees. To establish a comprehensive open-access dermatopathology dataset for educational, cross-referencing, and machine-learning purposes, we employed a hybrid workflow to curate and categorize images from the PubMed Central (PMC) repository. We used specific keywords to extract relevant images, and classified them using a novel hybrid method that combined deep learning-based image modality classification with figure caption analyses. Validation on 651 manually annotated images demonstrated the robustness of our workflow, with an F-score of 89.6\% for the deep learning approach, 61.0\% for the keyword-based retrieval method, and 90.4\% for the hybrid approach. We retrieved over 7,772 images across 166 diagnoses and released this fully annotated dataset, reviewed by board-certified dermatopathologists. Using our dataset as a challenging task, we found the current image analysis algorithm from OpenAI inadequate for analyzing dermatopathology images. In conclusion, we have developed a large, peer-reviewed, open-access dermatopathology image dataset, DermpathNet, which features a semi-automated curation workflow.
FinCriticalED: A Visual Benchmark for Financial Fact-Level OCR Evaluation
Nov 19, 2025Abstract:We introduce FinCriticalED (Financial Critical Error Detection), a visual benchmark for evaluating OCR and vision language models on financial documents at the fact level. Financial documents contain visually dense and table heavy layouts where numerical and temporal information is tightly coupled with structure. In high stakes settings, small OCR mistakes such as sign inversion or shifted dates can lead to materially different interpretations, while traditional OCR metrics like ROUGE and edit distance capture only surface level text similarity. \ficriticaled provides 500 image-HTML pairs with expert annotated financial facts covering over seven hundred numerical and temporal facts. It introduces three key contributions. First, it establishes the first fact level evaluation benchmark for financial document understanding, shifting evaluation from lexical overlap to domain critical factual correctness. Second, all annotations are created and verified by financial experts with strict quality control over signs, magnitudes, and temporal expressions. Third, we develop an LLM-as-Judge evaluation pipeline that performs structured fact extraction and contextual verification for visually complex financial documents. We benchmark OCR systems, open source vision language models, and proprietary models on FinCriticalED. Results show that although the strongest proprietary models achieve the highest factual accuracy, substantial errors remain in visually intricate numerical and temporal contexts. Through quantitative evaluation and expert case studies, FinCriticalED provides a rigorous foundation for advancing visual factual precision in financial and other precision critical domains.
MeCaMIL: Causality-Aware Multiple Instance Learning for Fair and Interpretable Whole Slide Image Diagnosis
Nov 14, 2025Abstract:Multiple instance learning (MIL) has emerged as the dominant paradigm for whole slide image (WSI) analysis in computational pathology, achieving strong diagnostic performance through patch-level feature aggregation. However, existing MIL methods face critical limitations: (1) they rely on attention mechanisms that lack causal interpretability, and (2) they fail to integrate patient demographics (age, gender, race), leading to fairness concerns across diverse populations. These shortcomings hinder clinical translation, where algorithmic bias can exacerbate health disparities. We introduce \textbf{MeCaMIL}, a causality-aware MIL framework that explicitly models demographic confounders through structured causal graphs. Unlike prior approaches treating demographics as auxiliary features, MeCaMIL employs principled causal inference -- leveraging do-calculus and collider structures -- to disentangle disease-relevant signals from spurious demographic correlations. Extensive evaluation on three benchmarks demonstrates state-of-the-art performance across CAMELYON16 (ACC/AUC/F1: 0.939/0.983/0.946), TCGA-Lung (0.935/0.979/0.931), and TCGA-Multi (0.977/0.993/0.970, five cancer types). Critically, MeCaMIL achieves superior fairness -- demographic disparity variance drops by over 65% relative reduction on average across attributes, with notable improvements for underserved populations. The framework generalizes to survival prediction (mean C-index: 0.653, +0.017 over best baseline across five cancer types). Ablation studies confirm causal graph structure is essential -- alternative designs yield 0.048 lower accuracy and 4.2x times worse fairness. These results establish MeCaMIL as a principled framework for fair, interpretable, and clinically actionable AI in digital pathology. Code will be released upon acceptance.
Two-Stage Decoupling Framework for Variable-Length Glaucoma Prognosis
Sep 15, 2025Abstract:Glaucoma is one of the leading causes of irreversible blindness worldwide. Glaucoma prognosis is essential for identifying at-risk patients and enabling timely intervention to prevent blindness. Many existing approaches rely on historical sequential data but are constrained by fixed-length inputs, limiting their flexibility. Additionally, traditional glaucoma prognosis methods often employ end-to-end models, which struggle with the limited size of glaucoma datasets. To address these challenges, we propose a Two-Stage Decoupling Framework (TSDF) for variable-length glaucoma prognosis. In the first stage, we employ a feature representation module that leverages self-supervised learning to aggregate multiple glaucoma datasets for training, disregarding differences in their supervisory information. This approach enables datasets of varying sizes to learn better feature representations. In the second stage, we introduce a temporal aggregation module that incorporates an attention-based mechanism to process sequential inputs of varying lengths, ensuring flexible and efficient utilization of all available data. This design significantly enhances model performance while maintaining a compact parameter size. Extensive experiments on two benchmark glaucoma datasets:the Ocular Hypertension Treatment Study (OHTS) and the Glaucoma Real-world Appraisal Progression Ensemble (GRAPE),which differ significantly in scale and clinical settings,demonstrate the effectiveness and robustness of our approach.
Data-Efficient Biomedical In-Context Learning: A Diversity-Enhanced Submodular Perspective
Aug 11, 2025Abstract:Recent progress in large language models (LLMs) has leveraged their in-context learning (ICL) abilities to enable quick adaptation to unseen biomedical NLP tasks. By incorporating only a few input-output examples into prompts, LLMs can rapidly perform these new tasks. While the impact of these demonstrations on LLM performance has been extensively studied, most existing approaches prioritize representativeness over diversity when selecting examples from large corpora. To address this gap, we propose Dual-Div, a diversity-enhanced data-efficient framework for demonstration selection in biomedical ICL. Dual-Div employs a two-stage retrieval and ranking process: First, it identifies a limited set of candidate examples from a corpus by optimizing both representativeness and diversity (with optional annotation for unlabeled data). Second, it ranks these candidates against test queries to select the most relevant and non-redundant demonstrations. Evaluated on three biomedical NLP tasks (named entity recognition (NER), relation extraction (RE), and text classification (TC)) using LLaMA 3.1 and Qwen 2.5 for inference, along with three retrievers (BGE-Large, BMRetriever, MedCPT), Dual-Div consistently outperforms baselines-achieving up to 5% higher macro-F1 scores-while demonstrating robustness to prompt permutations and class imbalance. Our findings establish that diversity in initial retrieval is more critical than ranking-stage optimization, and limiting demonstrations to 3-5 examples maximizes performance efficiency.
CXR-LT 2024: A MICCAI challenge on long-tailed, multi-label, and zero-shot disease classification from chest X-ray
Jun 09, 2025Abstract:The CXR-LT series is a community-driven initiative designed to enhance lung disease classification using chest X-rays (CXR). It tackles challenges in open long-tailed lung disease classification and enhances the measurability of state-of-the-art techniques. The first event, CXR-LT 2023, aimed to achieve these goals by providing high-quality benchmark CXR data for model development and conducting comprehensive evaluations to identify ongoing issues impacting lung disease classification performance. Building on the success of CXR-LT 2023, the CXR-LT 2024 expands the dataset to 377,110 chest X-rays (CXRs) and 45 disease labels, including 19 new rare disease findings. It also introduces a new focus on zero-shot learning to address limitations identified in the previous event. Specifically, CXR-LT 2024 features three tasks: (i) long-tailed classification on a large, noisy test set, (ii) long-tailed classification on a manually annotated "gold standard" subset, and (iii) zero-shot generalization to five previously unseen disease findings. This paper provides an overview of CXR-LT 2024, detailing the data curation process and consolidating state-of-the-art solutions, including the use of multimodal models for rare disease detection, advanced generative approaches to handle noisy labels, and zero-shot learning strategies for unseen diseases. Additionally, the expanded dataset enhances disease coverage to better represent real-world clinical settings, offering a valuable resource for future research. By synthesizing the insights and innovations of participating teams, we aim to advance the development of clinically realistic and generalizable diagnostic models for chest radiography.
Uncertainty-Aware Large Language Models for Explainable Disease Diagnosis
May 06, 2025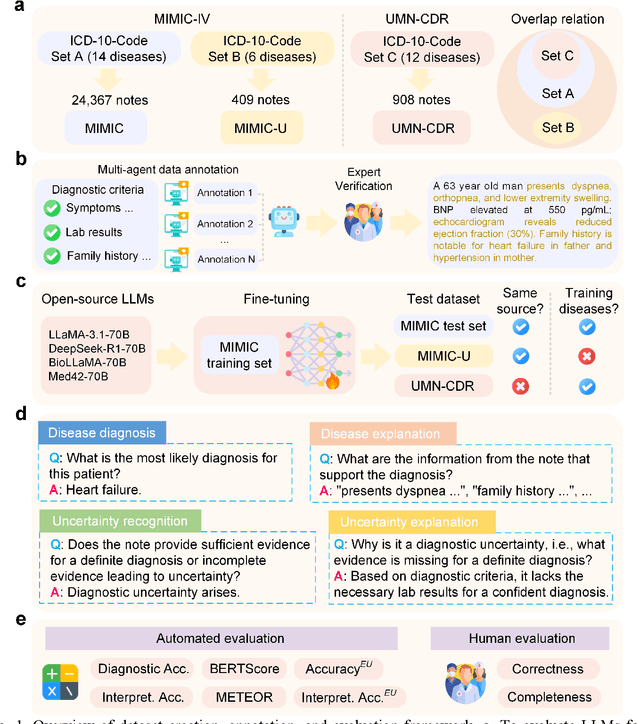
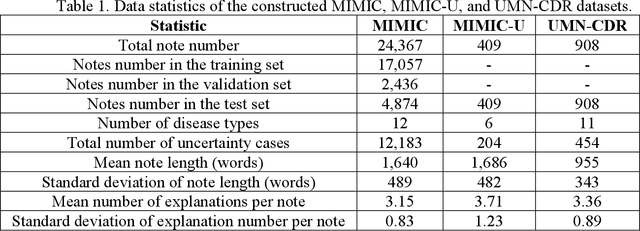
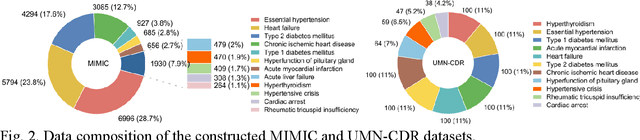
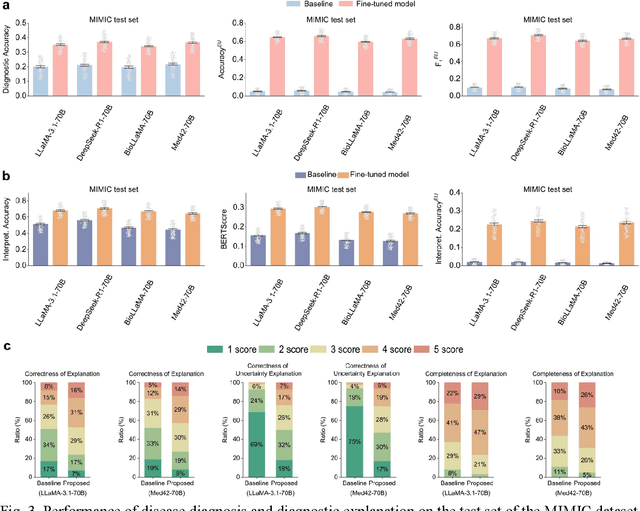
Abstract:Explainable disease diagnosis, which leverages patient information (e.g., signs and symptoms) and computational models to generate probable diagnoses and reasonings, offers clear clinical values. However, when clinical notes encompass insufficient evidence for a definite diagnosis, such as the absence of definitive symptoms, diagnostic uncertainty usually arises, increasing the risk of misdiagnosis and adverse outcomes. Although explicitly identifying and explaining diagnostic uncertainties is essential for trustworthy diagnostic systems, it remains under-explored. To fill this gap, we introduce ConfiDx, an uncertainty-aware large language model (LLM) created by fine-tuning open-source LLMs with diagnostic criteria. We formalized the task and assembled richly annotated datasets that capture varying degrees of diagnostic ambiguity. Evaluating ConfiDx on real-world datasets demonstrated that it excelled in identifying diagnostic uncertainties, achieving superior diagnostic performance, and generating trustworthy explanations for diagnoses and uncertainties. To our knowledge, this is the first study to jointly address diagnostic uncertainty recognition and explanation, substantially enhancing the reliability of automatic diagnostic systems.
FLAG-Trader: Fusion LLM-Agent with Gradient-based Reinforcement Learning for Financial Trading
Feb 19, 2025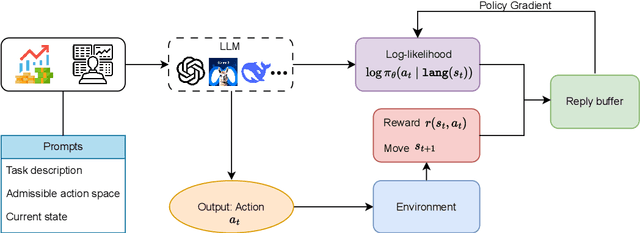
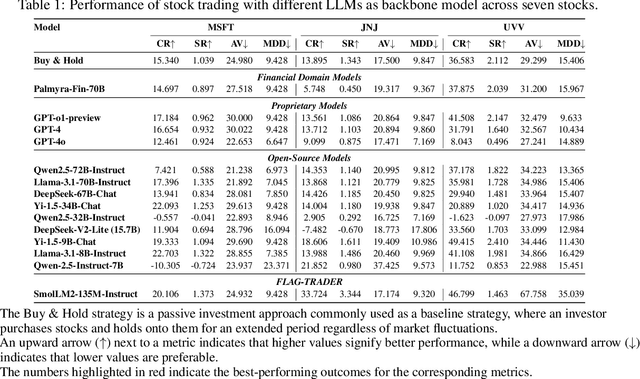
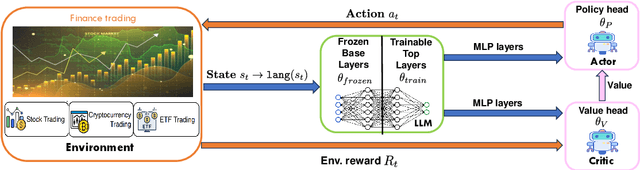
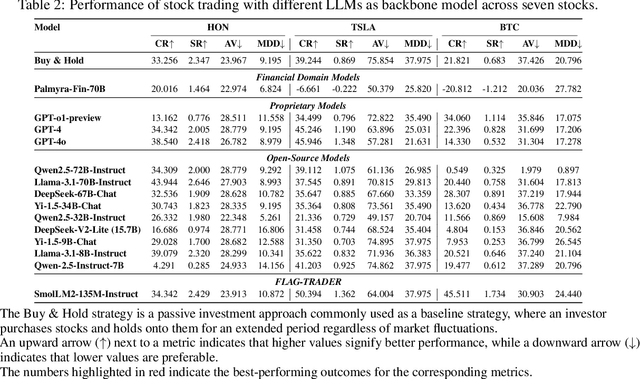
Abstract:Large language models (LLMs) fine-tuned on multimodal financial data have demonstrated impressive reasoning capabilities in various financial tasks. However, they often struggle with multi-step, goal-oriented scenarios in interactive financial markets, such as trading, where complex agentic approaches are required to improve decision-making. To address this, we propose \textsc{FLAG-Trader}, a unified architecture integrating linguistic processing (via LLMs) with gradient-driven reinforcement learning (RL) policy optimization, in which a partially fine-tuned LLM acts as the policy network, leveraging pre-trained knowledge while adapting to the financial domain through parameter-efficient fine-tuning. Through policy gradient optimization driven by trading rewards, our framework not only enhances LLM performance in trading but also improves results on other financial-domain tasks. We present extensive empirical evidence to validate these enhancements.
Continually Evolved Multimodal Foundation Models for Cancer Prognosis
Jan 30, 2025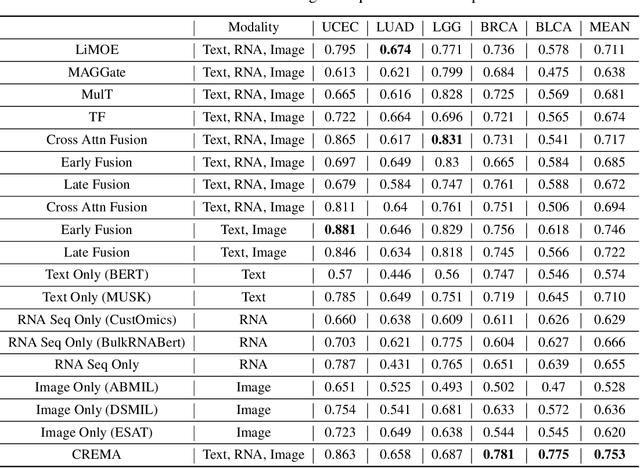
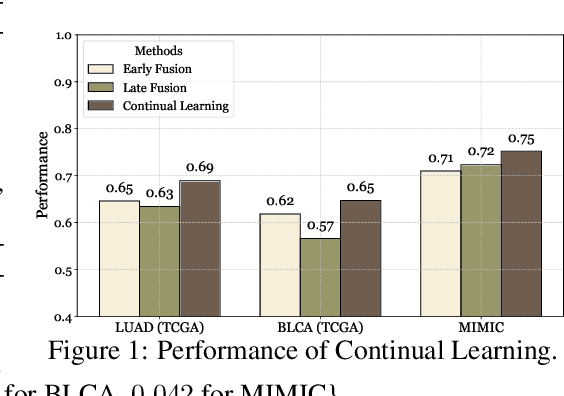
Abstract:Cancer prognosis is a critical task that involves predicting patient outcomes and survival rates. To enhance prediction accuracy, previous studies have integrated diverse data modalities, such as clinical notes, medical images, and genomic data, leveraging their complementary information. However, existing approaches face two major limitations. First, they struggle to incorporate newly arrived data with varying distributions into training, such as patient records from different hospitals, thus rendering sub-optimal generalizability and limited utility in real-world applications. Second, most multimodal integration methods rely on simplistic concatenation or task-specific pipelines, which fail to capture the complex interdependencies across modalities. To address these, we propose a continually evolving multi-modal foundation model. Extensive experiments on the TCGA dataset demonstrate the effectiveness of our approach, highlighting its potential to advance cancer prognosis by enabling robust and adaptive multimodal integration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge