Kaleb E Smith
FLAG-Trader: Fusion LLM-Agent with Gradient-based Reinforcement Learning for Financial Trading
Feb 19, 2025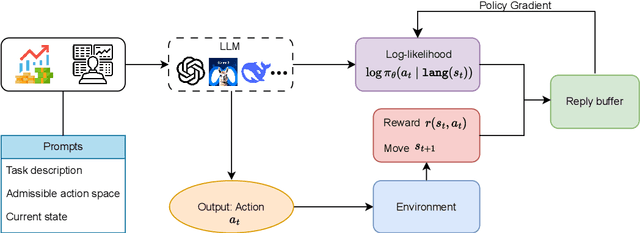
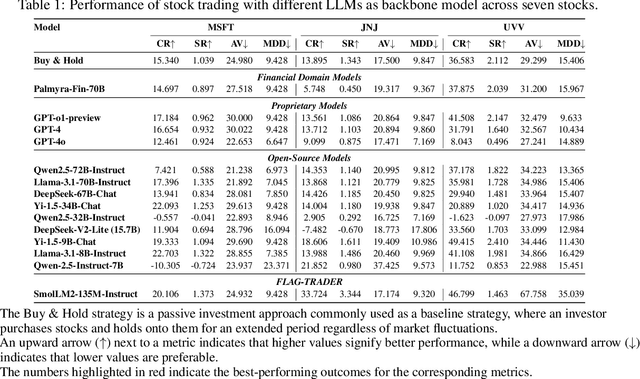
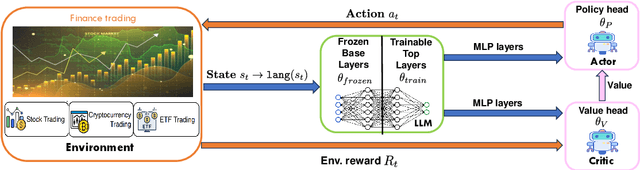
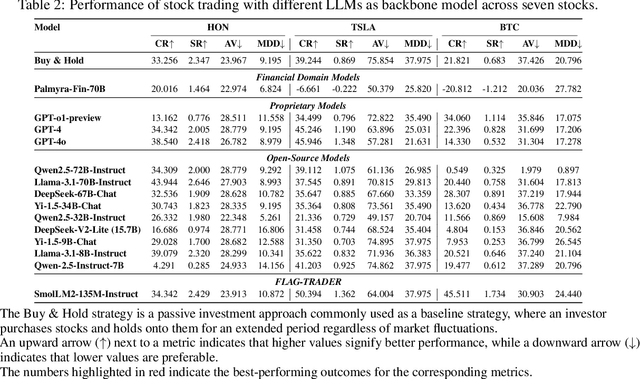
Abstract:Large language models (LLMs) fine-tuned on multimodal financial data have demonstrated impressive reasoning capabilities in various financial tasks. However, they often struggle with multi-step, goal-oriented scenarios in interactive financial markets, such as trading, where complex agentic approaches are required to improve decision-making. To address this, we propose \textsc{FLAG-Trader}, a unified architecture integrating linguistic processing (via LLMs) with gradient-driven reinforcement learning (RL) policy optimization, in which a partially fine-tuned LLM acts as the policy network, leveraging pre-trained knowledge while adapting to the financial domain through parameter-efficient fine-tuning. Through policy gradient optimization driven by trading rewards, our framework not only enhances LLM performance in trading but also improves results on other financial-domain tasks. We present extensive empirical evidence to validate these enhancements.
CLUE: A Clinical Language Understanding Evaluation for LLMs
Apr 11, 2024Abstract:Large Language Models (LLMs) have shown the potential to significantly contribute to patient care, diagnostics, and administrative processes. Emerging biomedical LLMs address healthcare-specific challenges, including privacy demands and computational constraints. However, evaluation of these models has primarily been limited to non-clinical tasks, which do not reflect the complexity of practical clinical applications. Additionally, there has been no thorough comparison between biomedical and general-domain LLMs for clinical tasks. To fill this gap, we present the Clinical Language Understanding Evaluation (CLUE), a benchmark tailored to evaluate LLMs on real-world clinical tasks. CLUE includes two novel datasets derived from MIMIC IV discharge letters and four existing tasks designed to test the practical applicability of LLMs in healthcare settings. Our evaluation covers several biomedical and general domain LLMs, providing insights into their clinical performance and applicability. CLUE represents a step towards a standardized approach to evaluating and developing LLMs in healthcare to align future model development with the real-world needs of clinical application. We publish our evaluation and data generation scripts: https://github.com/TIO-IKIM/CLUE.
Improving Generalizability of Extracting Social Determinants of Health Using Large Language Models through Prompt-tuning
Mar 19, 2024


Abstract:The progress in natural language processing (NLP) using large language models (LLMs) has greatly improved patient information extraction from clinical narratives. However, most methods based on the fine-tuning strategy have limited transfer learning ability for cross-domain applications. This study proposed a novel approach that employs a soft prompt-based learning architecture, which introduces trainable prompts to guide LLMs toward desired outputs. We examined two types of LLM architectures, including encoder-only GatorTron and decoder-only GatorTronGPT, and evaluated their performance for the extraction of social determinants of health (SDoH) using a cross-institution dataset from the 2022 n2c2 challenge and a cross-disease dataset from the University of Florida (UF) Health. The results show that decoder-only LLMs with prompt tuning achieved better performance in cross-domain applications. GatorTronGPT achieved the best F1 scores for both datasets, outperforming traditional fine-tuned GatorTron by 8.9% and 21.8% in a cross-institution setting, and 5.5% and 14.5% in a cross-disease setting.
Generative Large Language Models Are All-purpose Text Analytics Engines: Text-to-text Learning Is All Your Need
Dec 11, 2023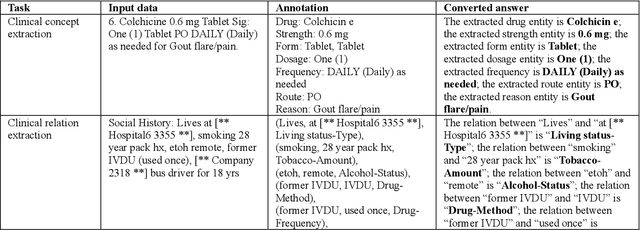
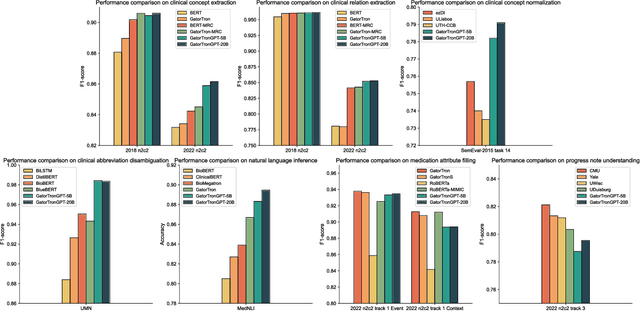
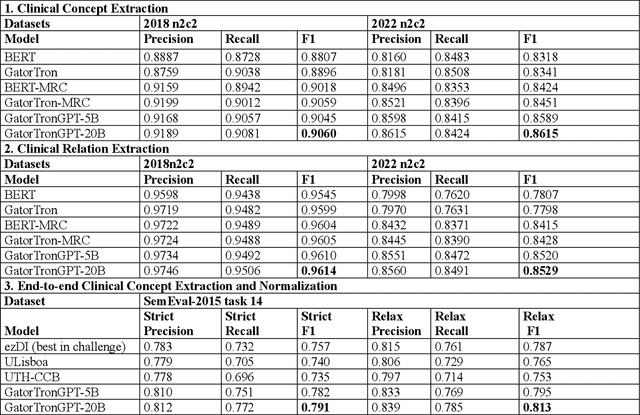
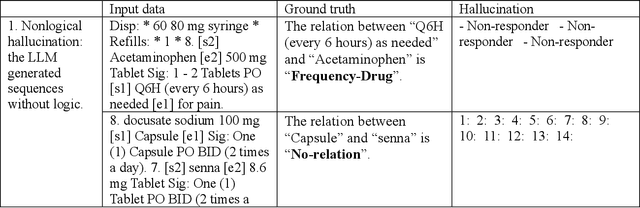
Abstract:Objective To solve major clinical natural language processing (NLP) tasks using a unified text-to-text learning architecture based on a generative large language model (LLM) via prompt tuning. Methods We formulated 7 key clinical NLP tasks as text-to-text learning and solved them using one unified generative clinical LLM, GatorTronGPT, developed using GPT-3 architecture and trained with up to 20 billion parameters. We adopted soft prompts (i.e., trainable vectors) with frozen LLM, where the LLM parameters were not updated (i.e., frozen) and only the vectors of soft prompts were updated, known as prompt tuning. We added additional soft prompts as a prefix to the input layer, which were optimized during the prompt tuning. We evaluated the proposed method using 7 clinical NLP tasks and compared them with previous task-specific solutions based on Transformer models. Results and Conclusion The proposed approach achieved state-of-the-art performance for 5 out of 7 major clinical NLP tasks using one unified generative LLM. Our approach outperformed previous task-specific transformer models by ~3% for concept extraction and 7% for relation extraction applied to social determinants of health, 3.4% for clinical concept normalization, 3.4~10% for clinical abbreviation disambiguation, and 5.5~9% for natural language inference. Our approach also outperformed a previously developed prompt-based machine reading comprehension (MRC) model, GatorTron-MRC, for clinical concept and relation extraction. The proposed approach can deliver the ``one model for all`` promise from training to deployment using a unified generative LLM.
On the Impact of Cross-Domain Data on German Language Models
Oct 13, 2023Abstract:Traditionally, large language models have been either trained on general web crawls or domain-specific data. However, recent successes of generative large language models, have shed light on the benefits of cross-domain datasets. To examine the significance of prioritizing data diversity over quality, we present a German dataset comprising texts from five domains, along with another dataset aimed at containing high-quality data. Through training a series of models ranging between 122M and 750M parameters on both datasets, we conduct a comprehensive benchmark on multiple downstream tasks. Our findings demonstrate that the models trained on the cross-domain dataset outperform those trained on quality data alone, leading to improvements up to $4.45\%$ over the previous state-of-the-art. The models are available at https://huggingface.co/ikim-uk-essen
Model Tuning or Prompt Tuning? A Study of Large Language Models for Clinical Concept and Relation Extraction
Oct 10, 2023Abstract:Objective To develop soft prompt-based learning algorithms for large language models (LLMs), examine the shape of prompts, prompt-tuning using frozen/unfrozen LLMs, transfer learning, and few-shot learning abilities. Methods We developed a soft prompt-based LLM model and compared 4 training strategies including (1) fine-tuning without prompts; (2) hard-prompt with unfrozen LLMs; (3) soft-prompt with unfrozen LLMs; and (4) soft-prompt with frozen LLMs. We evaluated 7 pretrained LLMs using the 4 training strategies for clinical concept and relation extraction on two benchmark datasets. We evaluated the transfer learning ability of the prompt-based learning algorithms in a cross-institution setting. We also assessed the few-shot learning ability. Results and Conclusion When LLMs are unfrozen, GatorTron-3.9B with soft prompting achieves the best strict F1-scores of 0.9118 and 0.8604 for concept extraction, outperforming the traditional fine-tuning and hard prompt-based models by 0.6~3.1% and 1.2~2.9%, respectively; GatorTron-345M with soft prompting achieves the best F1-scores of 0.8332 and 0.7488 for end-to-end relation extraction, outperforming the other two models by 0.2~2% and 0.6~11.7%, respectively. When LLMs are frozen, small (i.e., 345 million parameters) LLMs have a big gap to be competitive with unfrozen models; scaling LLMs up to billions of parameters makes frozen LLMs competitive with unfrozen LLMs. For cross-institute evaluation, soft prompting with a frozen GatorTron-8.9B model achieved the best performance. This study demonstrates that (1) machines can learn soft prompts better than humans, (2) frozen LLMs have better few-shot learning ability and transfer learning ability to facilitate muti-institution applications, and (3) frozen LLMs require large models.
A Study of Generative Large Language Model for Medical Research and Healthcare
May 22, 2023Abstract:There is enormous enthusiasm and concerns in using large language models (LLMs) in healthcare, yet current assumptions are all based on general-purpose LLMs such as ChatGPT. This study develops a clinical generative LLM, GatorTronGPT, using 277 billion words of mixed clinical and English text with a GPT-3 architecture of 20 billion parameters. GatorTronGPT improves biomedical natural language processing for medical research. Synthetic NLP models trained using GatorTronGPT generated text outperform NLP models trained using real-world clinical text. Physicians Turing test using 1 (worst) to 9 (best) scale shows that there is no significant difference in linguistic readability (p = 0.22; 6.57 of GatorTronGPT compared with 6.93 of human) and clinical relevance (p = 0.91; 7.0 of GatorTronGPT compared with 6.97 of human) and that physicians cannot differentiate them (p < 0.001). This study provides insights on the opportunities and challenges of LLMs for medical research and healthcare.
Conditional GAN for timeseries generation
Jun 30, 2020



Abstract:It is abundantly clear that time dependent data is a vital source of information in the world. The challenge has been for applications in machine learning to gain access to a considerable amount of quality data needed for algorithm development and analysis. Modeling synthetic data using a Generative Adversarial Network (GAN) has been at the heart of providing a viable solution. Our work focuses on one dimensional times series and explores the few shot approach, which is the ability of an algorithm to perform well with limited data. This work attempts to ease the frustration by proposing a new architecture, Time Series GAN (TSGAN), to model realistic time series data. We evaluate TSGAN on 70 data sets from a benchmark time series database. Our results demonstrate that TSGAN performs better than the competition both quantitatively using the Frechet Inception Score (FID) metric, and qualitatively when classification is used as the evaluation criteria.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge