Aokun Chen
UF-HOBI at "Discharge Me!": A Hybrid Solution for Discharge Summary Generation Through Prompt-based Tuning of GatorTronGPT Models
Jul 22, 2024



Abstract:Automatic generation of discharge summaries presents significant challenges due to the length of clinical documentation, the dispersed nature of patient information, and the diverse terminology used in healthcare. This paper presents a hybrid solution for generating discharge summary sections as part of our participation in the "Discharge Me!" Challenge at the BioNLP 2024 Shared Task. We developed a two-stage generation method using both extractive and abstractive techniques, in which we first apply name entity recognition (NER) to extract key clinical concepts, which are then used as input for a prompt-tuning-based GatorTronGPT model to generate coherent text for two important sections including "Brief Hospital Course" and "Discharge Instructions". Our system was ranked 5th in this challenge, achieving an overall score of 0.284. The results demonstrate the effectiveness of our hybrid solution in improving the quality of automated discharge section generation.
* BIONLP 2024 and Shared Tasks @ ACL 2024
Me LLaMA: Foundation Large Language Models for Medical Applications
Feb 20, 2024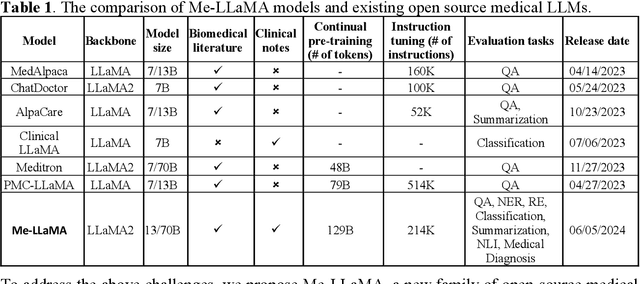



Abstract:Recent large language models (LLMs) like ChatGPT and LLaMA have shown great promise in many AI applications. However, their performance on medical tasks is suboptimal and can be further improved by training on large domain-specific datasets. This study introduces Me LLaMA, a medical LLM family including foundation models - Me LLaMA 13/70B and their chat-enhanced versions - Me LLaMA 13/70B-chat, developed through the continual pre-training and instruction tuning of LLaMA2 using large medical data. Our domain-specific data suite for training and evaluation, includes a large-scale continual pre-training dataset with 129B tokens, an instruction tuning dataset with 214k samples, and a medical evaluation benchmark (MIBE) across six tasks with 14 datasets. Our extensive evaluation using MIBE shows that Me LLaMA models surpass existing open-source medical LLMs in zero-shot and few-shot learning and outperform commercial giants like ChatGPT on 6 out of 8 datasets and GPT-4 in 3 out of 8 datasets. In addition, we empirically investigated the catastrophic forgetting problem, and our results show that Me LLaMA models outperform other medical LLMs. Me LLaMA is one of the first and largest open-source foundational LLMs designed for the medical domain, using both biomedical and clinical data. It exhibits superior performance across both general and medical tasks compared to other medical LLMs, rendering it an attractive choice for medical AI applications. All resources are available at: https://github.com/BIDS-Xu-Lab/Me-LLaMA.
Generative Large Language Models Are All-purpose Text Analytics Engines: Text-to-text Learning Is All Your Need
Dec 11, 2023
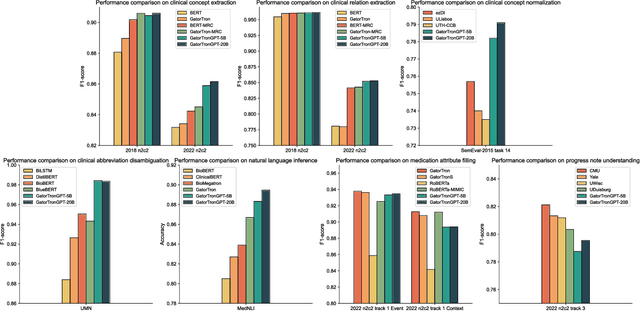
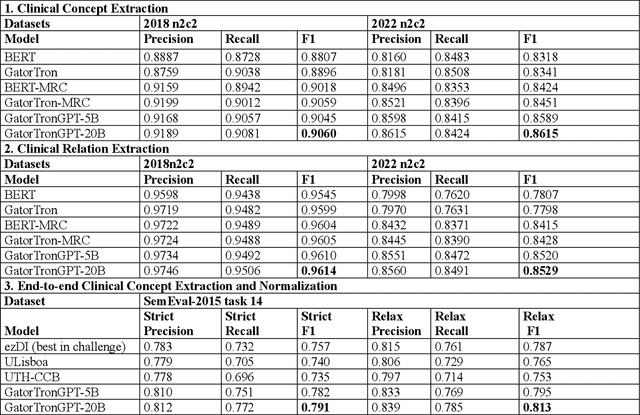
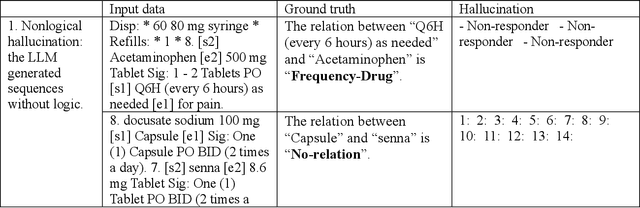
Abstract:Objective To solve major clinical natural language processing (NLP) tasks using a unified text-to-text learning architecture based on a generative large language model (LLM) via prompt tuning. Methods We formulated 7 key clinical NLP tasks as text-to-text learning and solved them using one unified generative clinical LLM, GatorTronGPT, developed using GPT-3 architecture and trained with up to 20 billion parameters. We adopted soft prompts (i.e., trainable vectors) with frozen LLM, where the LLM parameters were not updated (i.e., frozen) and only the vectors of soft prompts were updated, known as prompt tuning. We added additional soft prompts as a prefix to the input layer, which were optimized during the prompt tuning. We evaluated the proposed method using 7 clinical NLP tasks and compared them with previous task-specific solutions based on Transformer models. Results and Conclusion The proposed approach achieved state-of-the-art performance for 5 out of 7 major clinical NLP tasks using one unified generative LLM. Our approach outperformed previous task-specific transformer models by ~3% for concept extraction and 7% for relation extraction applied to social determinants of health, 3.4% for clinical concept normalization, 3.4~10% for clinical abbreviation disambiguation, and 5.5~9% for natural language inference. Our approach also outperformed a previously developed prompt-based machine reading comprehension (MRC) model, GatorTron-MRC, for clinical concept and relation extraction. The proposed approach can deliver the ``one model for all`` promise from training to deployment using a unified generative LLM.
On the Impact of Cross-Domain Data on German Language Models
Oct 13, 2023Abstract:Traditionally, large language models have been either trained on general web crawls or domain-specific data. However, recent successes of generative large language models, have shed light on the benefits of cross-domain datasets. To examine the significance of prioritizing data diversity over quality, we present a German dataset comprising texts from five domains, along with another dataset aimed at containing high-quality data. Through training a series of models ranging between 122M and 750M parameters on both datasets, we conduct a comprehensive benchmark on multiple downstream tasks. Our findings demonstrate that the models trained on the cross-domain dataset outperform those trained on quality data alone, leading to improvements up to $4.45\%$ over the previous state-of-the-art. The models are available at https://huggingface.co/ikim-uk-essen
Model Tuning or Prompt Tuning? A Study of Large Language Models for Clinical Concept and Relation Extraction
Oct 10, 2023Abstract:Objective To develop soft prompt-based learning algorithms for large language models (LLMs), examine the shape of prompts, prompt-tuning using frozen/unfrozen LLMs, transfer learning, and few-shot learning abilities. Methods We developed a soft prompt-based LLM model and compared 4 training strategies including (1) fine-tuning without prompts; (2) hard-prompt with unfrozen LLMs; (3) soft-prompt with unfrozen LLMs; and (4) soft-prompt with frozen LLMs. We evaluated 7 pretrained LLMs using the 4 training strategies for clinical concept and relation extraction on two benchmark datasets. We evaluated the transfer learning ability of the prompt-based learning algorithms in a cross-institution setting. We also assessed the few-shot learning ability. Results and Conclusion When LLMs are unfrozen, GatorTron-3.9B with soft prompting achieves the best strict F1-scores of 0.9118 and 0.8604 for concept extraction, outperforming the traditional fine-tuning and hard prompt-based models by 0.6~3.1% and 1.2~2.9%, respectively; GatorTron-345M with soft prompting achieves the best F1-scores of 0.8332 and 0.7488 for end-to-end relation extraction, outperforming the other two models by 0.2~2% and 0.6~11.7%, respectively. When LLMs are frozen, small (i.e., 345 million parameters) LLMs have a big gap to be competitive with unfrozen models; scaling LLMs up to billions of parameters makes frozen LLMs competitive with unfrozen LLMs. For cross-institute evaluation, soft prompting with a frozen GatorTron-8.9B model achieved the best performance. This study demonstrates that (1) machines can learn soft prompts better than humans, (2) frozen LLMs have better few-shot learning ability and transfer learning ability to facilitate muti-institution applications, and (3) frozen LLMs require large models.
A Study of Generative Large Language Model for Medical Research and Healthcare
May 22, 2023Abstract:There is enormous enthusiasm and concerns in using large language models (LLMs) in healthcare, yet current assumptions are all based on general-purpose LLMs such as ChatGPT. This study develops a clinical generative LLM, GatorTronGPT, using 277 billion words of mixed clinical and English text with a GPT-3 architecture of 20 billion parameters. GatorTronGPT improves biomedical natural language processing for medical research. Synthetic NLP models trained using GatorTronGPT generated text outperform NLP models trained using real-world clinical text. Physicians Turing test using 1 (worst) to 9 (best) scale shows that there is no significant difference in linguistic readability (p = 0.22; 6.57 of GatorTronGPT compared with 6.93 of human) and clinical relevance (p = 0.91; 7.0 of GatorTronGPT compared with 6.97 of human) and that physicians cannot differentiate them (p < 0.001). This study provides insights on the opportunities and challenges of LLMs for medical research and healthcare.
Identifying Symptoms of Delirium from Clinical Narratives Using Natural Language Processing
Mar 31, 2023



Abstract:Delirium is an acute decline or fluctuation in attention, awareness, or other cognitive function that can lead to serious adverse outcomes. Despite the severe outcomes, delirium is frequently unrecognized and uncoded in patients' electronic health records (EHRs) due to its transient and diverse nature. Natural language processing (NLP), a key technology that extracts medical concepts from clinical narratives, has shown great potential in studies of delirium outcomes and symptoms. To assist in the diagnosis and phenotyping of delirium, we formed an expert panel to categorize diverse delirium symptoms, composed annotation guidelines, created a delirium corpus with diverse delirium symptoms, and developed NLP methods to extract delirium symptoms from clinical notes. We compared 5 state-of-the-art transformer models including 2 models (BERT and RoBERTa) from the general domain and 3 models (BERT_MIMIC, RoBERTa_MIMIC, and GatorTron) from the clinical domain. GatorTron achieved the best strict and lenient F1 scores of 0.8055 and 0.8759, respectively. We conducted an error analysis to identify challenges in annotating delirium symptoms and developing NLP systems. To the best of our knowledge, this is the first large language model-based delirium symptom extraction system. Our study lays the foundation for the future development of computable phenotypes and diagnosis methods for delirium.
Contextualized Medication Information Extraction Using Transformer-based Deep Learning Architectures
Mar 14, 2023Abstract:Objective: To develop a natural language processing (NLP) system to extract medications and contextual information that help understand drug changes. This project is part of the 2022 n2c2 challenge. Materials and methods: We developed NLP systems for medication mention extraction, event classification (indicating medication changes discussed or not), and context classification to classify medication changes context into 5 orthogonal dimensions related to drug changes. We explored 6 state-of-the-art pretrained transformer models for the three subtasks, including GatorTron, a large language model pretrained using >90 billion words of text (including >80 billion words from >290 million clinical notes identified at the University of Florida Health). We evaluated our NLP systems using annotated data and evaluation scripts provided by the 2022 n2c2 organizers. Results:Our GatorTron models achieved the best F1-scores of 0.9828 for medication extraction (ranked 3rd), 0.9379 for event classification (ranked 2nd), and the best micro-average accuracy of 0.9126 for context classification. GatorTron outperformed existing transformer models pretrained using smaller general English text and clinical text corpora, indicating the advantage of large language models. Conclusion: This study demonstrated the advantage of using large transformer models for contextual medication information extraction from clinical narratives.
Computer Users Have Unique Yet Temporally Inconsistent Computer Usage Profiles
May 20, 2021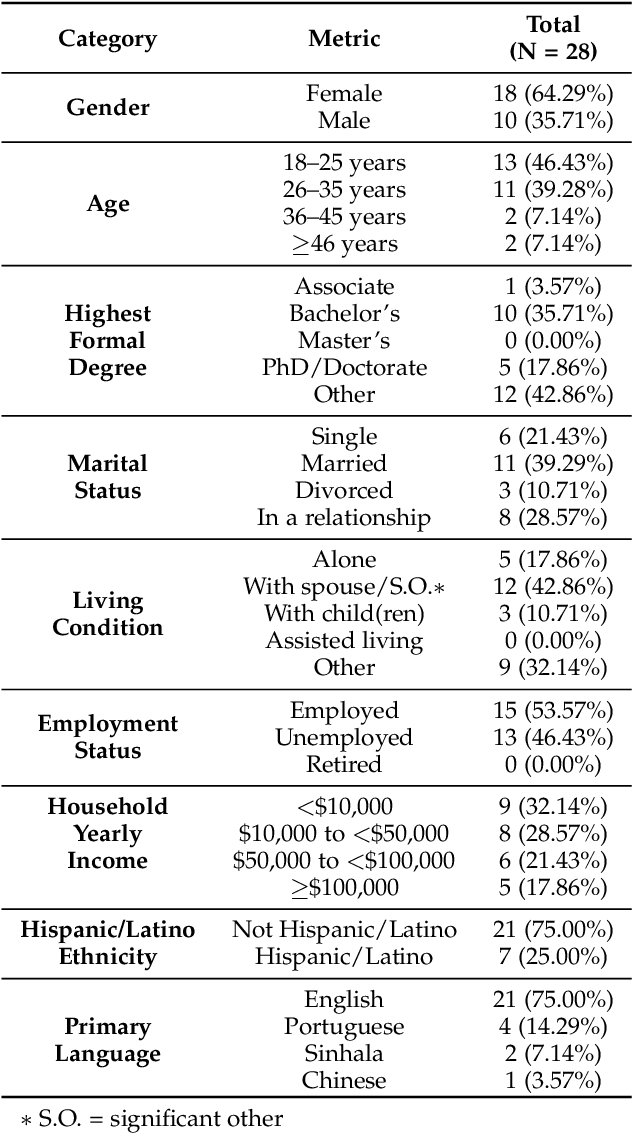
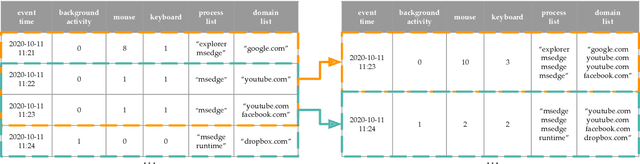
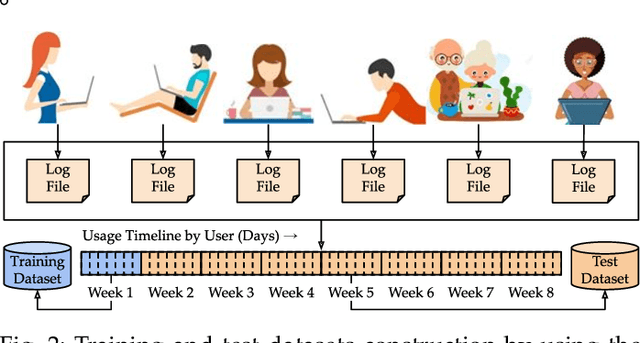
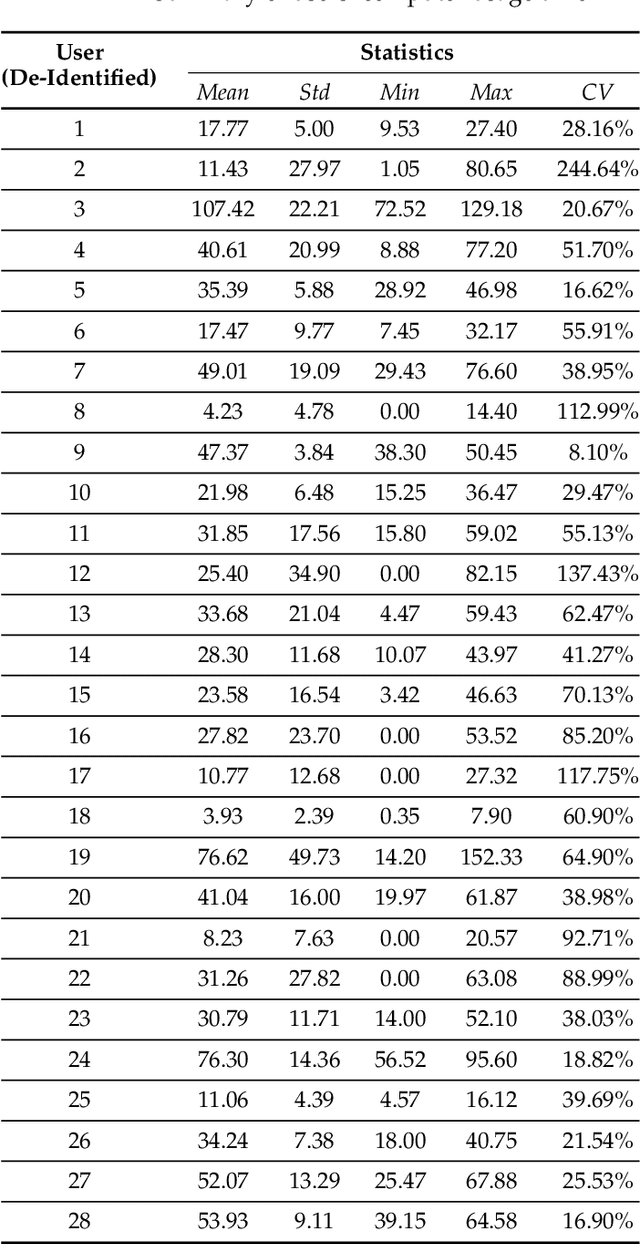
Abstract:This paper investigates whether computer usage profiles comprised of process-, network-, mouse- and keystroke-related events are unique and temporally consistent in a naturalistic setting, discussing challenges and opportunities of using such profiles in applications of continuous authentication. We collected ecologically-valid computer usage profiles from 28 MS Windows 10 computer users over 8 weeks and submitted this data to comprehensive machine learning analysis involving a diverse set of online and offline classifiers. We found that (i) computer usage profiles have the potential to uniquely characterize computer users (with a maximum F-score of 99.94%); (ii) network-related events were the most useful features to properly recognize profiles (95.14% of the top features distinguishing users being network-related); (iii) user profiles were mostly inconsistent over the 8-week data collection period, with 92.86% of users exhibiting drifts in terms of time and usage habits; and (iv) online models are better suited to handle computer usage profiles compared to offline models (maximum F-score for each approach was 95.99% and 99.94%, respectively).
Learning Fast and Slow: PROPEDEUTICA for Real-time Malware Detection
Dec 04, 2017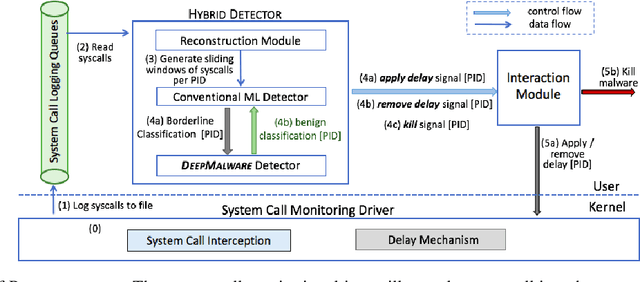
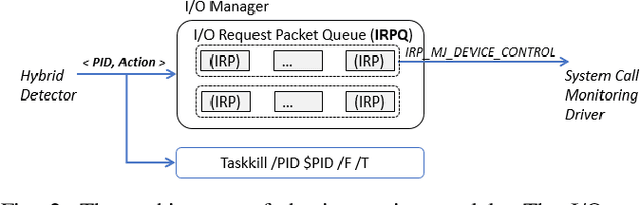
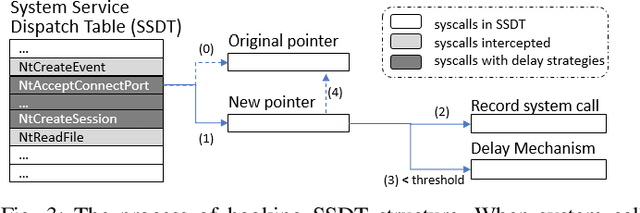
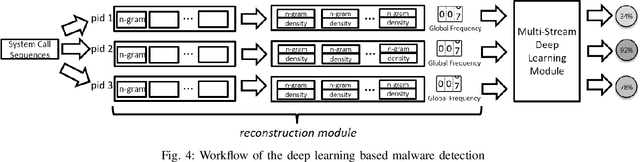
Abstract:In this paper, we introduce and evaluate PROPEDEUTICA, a novel methodology and framework for efficient and effective real-time malware detection, leveraging the best of conventional machine learning (ML) and deep learning (DL) algorithms. In PROPEDEUTICA, all software processes in the system start execution subjected to a conventional ML detector for fast classification. If a piece of software receives a borderline classification, it is subjected to further analysis via more performance expensive and more accurate DL methods, via our newly proposed DL algorithm DEEPMALWARE. Further, we introduce delays to the execution of software subjected to deep learning analysis as a way to "buy time" for DL analysis and to rate-limit the impact of possible malware in the system. We evaluated PROPEDEUTICA with a set of 9,115 malware samples and 877 commonly used benign software samples from various categories for the Windows OS. Our results show that the false positive rate for conventional ML methods can reach 20%, and for modern DL methods it is usually below 6%. However, the classification time for DL can be 100X longer than conventional ML methods. PROPEDEUTICA improved the detection F1-score from 77.54% (conventional ML method) to 90.25%, and reduced the detection time by 54.86%. Further, the percentage of software subjected to DL analysis was approximately 40% on average. Further, the application of delays in software subjected to ML reduced the detection time by approximately 10%. Finally, we found and discussed a discrepancy between the detection accuracy offline (analysis after all traces are collected) and on-the-fly (analysis in tandem with trace collection). Our insights show that conventional ML and modern DL-based malware detectors in isolation cannot meet the needs of efficient and effective malware detection: high accuracy, low false positive rate, and short classification time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge