Christoph M. Friedrich
eTracer: Towards Traceable Text Generation via Claim-Level Grounding
Jan 07, 2026Abstract:How can system-generated responses be efficiently verified, especially in the high-stakes biomedical domain? To address this challenge, we introduce eTracer, a plug-and-play framework that enables traceable text generation by grounding claims against contextual evidence. Through post-hoc grounding, each response claim is aligned with contextual evidence that either supports or contradicts it. Building on claim-level grounding results, eTracer not only enables users to precisely trace responses back to their contextual source but also quantifies response faithfulness, thereby enabling the verifiability and trustworthiness of generated responses. Experiments show that our claim-level grounding approach alleviates the limitations of conventional grounding methods in aligning generated statements with contextual sentence-level evidence, resulting in substantial improvements in overall grounding quality and user verification efficiency. The code and data are available at https://github.com/chubohao/eTracer.
Evaluating Compliance with Visualization Guidelines in Diagrams for Scientific Publications Using Large Vision Language Models
Jun 24, 2025Abstract:Diagrams are widely used to visualize data in publications. The research field of data visualization deals with defining principles and guidelines for the creation and use of these diagrams, which are often not known or adhered to by researchers, leading to misinformation caused by providing inaccurate or incomplete information. In this work, large Vision Language Models (VLMs) are used to analyze diagrams in order to identify potential problems in regards to selected data visualization principles and guidelines. To determine the suitability of VLMs for these tasks, five open source VLMs and five prompting strategies are compared using a set of questions derived from selected data visualization guidelines. The results show that the employed VLMs work well to accurately analyze diagram types (F1-score 82.49 %), 3D effects (F1-score 98.55 %), axes labels (F1-score 76.74 %), lines (RMSE 1.16), colors (RMSE 1.60) and legends (F1-score 96.64 %, RMSE 0.70), while they cannot reliably provide feedback about the image quality (F1-score 0.74 %) and tick marks/labels (F1-score 46.13 %). Among the employed VLMs, Qwen2.5VL performs best, and the summarizing prompting strategy performs best for most of the experimental questions. It is shown that VLMs can be used to automatically identify a number of potential issues in diagrams, such as missing axes labels, missing legends, and unnecessary 3D effects. The approach laid out in this work can be extended for further aspects of data visualization.
A Multimodal Pipeline for Clinical Data Extraction: Applying Vision-Language Models to Scans of Transfusion Reaction Reports
Apr 28, 2025Abstract:Despite the growing adoption of electronic health records, many processes still rely on paper documents, reflecting the heterogeneous real-world conditions in which healthcare is delivered. The manual transcription process is time-consuming and prone to errors when transferring paper-based data to digital formats. To streamline this workflow, this study presents an open-source pipeline that extracts and categorizes checkbox data from scanned documents. Demonstrated on transfusion reaction reports, the design supports adaptation to other checkbox-rich document types. The proposed method integrates checkbox detection, multilingual optical character recognition (OCR) and multilingual vision-language models (VLMs). The pipeline achieves high precision and recall compared against annually compiled gold-standards from 2017 to 2024. The result is a reduction in administrative workload and accurate regulatory reporting. The open-source availability of this pipeline encourages self-hosted parsing of checkbox forms.
An Enhanced Harmonic Densely Connected Hybrid Transformer Network Architecture for Chronic Wound Segmentation Utilising Multi-Colour Space Tensor Merging
Oct 04, 2024



Abstract:Chronic wounds and associated complications present ever growing burdens for clinics and hospitals world wide. Venous, arterial, diabetic, and pressure wounds are becoming increasingly common globally. These conditions can result in highly debilitating repercussions for those affected, with limb amputations and increased mortality risk resulting from infection becoming more common. New methods to assist clinicians in chronic wound care are therefore vital to maintain high quality care standards. This paper presents an improved HarDNet segmentation architecture which integrates a contrast-eliminating component in the initial layers of the network to enhance feature learning. We also utilise a multi-colour space tensor merging process and adjust the harmonic shape of the convolution blocks to facilitate these additional features. We train our proposed model using wound images from light-skinned patients and test the model on two test sets (one set with ground truth, and one without) comprising only darker-skinned cases. Subjective ratings are obtained from clinical wound experts with intraclass correlation coefficient used to determine inter-rater reliability. For the dark-skin tone test set with ground truth, we demonstrate improvements in terms of Dice similarity coefficient (+0.1221) and intersection over union (+0.1274). Qualitative analysis showed high expert ratings, with improvements of >3% demonstrated when comparing the baseline model with the proposed model. This paper presents the first study to focus on darker-skin tones for chronic wound segmentation using models trained only on wound images exhibiting lighter skin. Diabetes is highly prevalent in countries where patients have darker skin tones, highlighting the need for a greater focus on such cases. Additionally, we conduct the largest qualitative study to date for chronic wound segmentation.
SALT: Introducing a Framework for Hierarchical Segmentations in Medical Imaging using Softmax for Arbitrary Label Trees
Jul 11, 2024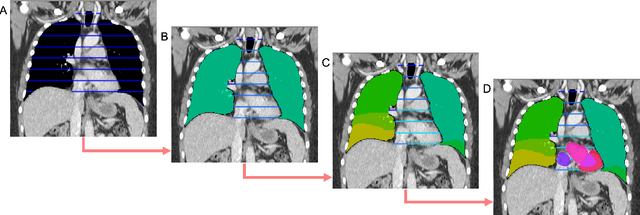
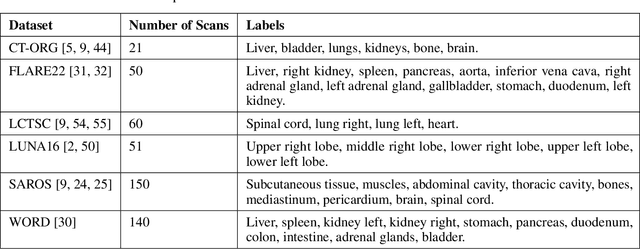
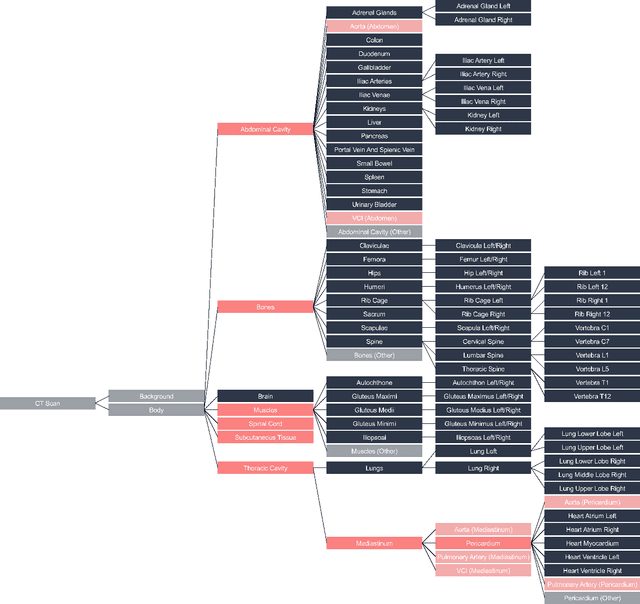
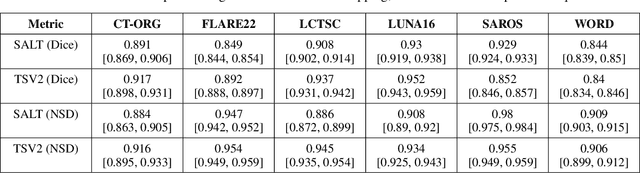
Abstract:Traditional segmentation networks approach anatomical structures as standalone elements, overlooking the intrinsic hierarchical connections among them. This study introduces Softmax for Arbitrary Label Trees (SALT), a novel approach designed to leverage the hierarchical relationships between labels, improving the efficiency and interpretability of the segmentations. This study introduces a novel segmentation technique for CT imaging, which leverages conditional probabilities to map the hierarchical structure of anatomical landmarks, such as the spine's division into lumbar, thoracic, and cervical regions and further into individual vertebrae. The model was developed using the SAROS dataset from The Cancer Imaging Archive (TCIA), comprising 900 body region segmentations from 883 patients. The dataset was further enhanced by generating additional segmentations with the TotalSegmentator, for a total of 113 labels. The model was trained on 600 scans, while validation and testing were conducted on 150 CT scans. Performance was assessed using the Dice score across various datasets, including SAROS, CT-ORG, FLARE22, LCTSC, LUNA16, and WORD. Among the evaluated datasets, SALT achieved its best results on the LUNA16 and SAROS datasets, with Dice scores of 0.93 and 0.929 respectively. The model demonstrated reliable accuracy across other datasets, scoring 0.891 on CT-ORG and 0.849 on FLARE22. The LCTSC dataset showed a score of 0.908 and the WORD dataset also showed good performance with a score of 0.844. SALT used the hierarchical structures inherent in the human body to achieve whole-body segmentations with an average of 35 seconds for 100 slices. This rapid processing underscores its potential for integration into clinical workflows, facilitating the automatic and efficient computation of full-body segmentations with each CT scan, thus enhancing diagnostic processes and patient care.
WisPerMed at BioLaySumm: Adapting Autoregressive Large Language Models for Lay Summarization of Scientific Articles
May 20, 2024Abstract:This paper details the efforts of the WisPerMed team in the BioLaySumm2024 Shared Task on automatic lay summarization in the biomedical domain, aimed at making scientific publications accessible to non-specialists. Large language models (LLMs), specifically the BioMistral and Llama3 models, were fine-tuned and employed to create lay summaries from complex scientific texts. The summarization performance was enhanced through various approaches, including instruction tuning, few-shot learning, and prompt variations tailored to incorporate specific context information. The experiments demonstrated that fine-tuning generally led to the best performance across most evaluated metrics. Few-shot learning notably improved the models' ability to generate relevant and factually accurate texts, particularly when using a well-crafted prompt. Additionally, a Dynamic Expert Selection (DES) mechanism to optimize the selection of text outputs based on readability and factuality metrics was developed. Out of 54 participants, the WisPerMed team reached the 4th place, measured by readability, factuality, and relevance. Determined by the overall score, our approach improved upon the baseline by approx. 5.5 percentage points and was only approx 1.5 percentage points behind the first place.
WisPerMed at "Discharge Me!": Advancing Text Generation in Healthcare with Large Language Models, Dynamic Expert Selection, and Priming Techniques on MIMIC-IV
May 18, 2024



Abstract:This study aims to leverage state of the art language models to automate generating the "Brief Hospital Course" and "Discharge Instructions" sections of Discharge Summaries from the MIMIC-IV dataset, reducing clinicians' administrative workload. We investigate how automation can improve documentation accuracy, alleviate clinician burnout, and enhance operational efficacy in healthcare facilities. This research was conducted within our participation in the Shared Task Discharge Me! at BioNLP @ ACL 2024. Various strategies were employed, including few-shot learning, instruction tuning, and Dynamic Expert Selection (DES), to develop models capable of generating the required text sections. Notably, utilizing an additional clinical domain-specific dataset demonstrated substantial potential to enhance clinical language processing. The DES method, which optimizes the selection of text outputs from multiple predictions, proved to be especially effective. It achieved the highest overall score of 0.332 in the competition, surpassing single-model outputs. This finding suggests that advanced deep learning methods in combination with DES can effectively automate parts of electronic health record documentation. These advancements could enhance patient care by freeing clinician time for patient interactions. The integration of text selection strategies represents a promising avenue for further research.
ROCOv2: Radiology Objects in COntext Version 2, an Updated Multimodal Image Dataset
May 16, 2024Abstract:Automated medical image analysis systems often require large amounts of training data with high quality labels, which are difficult and time consuming to generate. This paper introduces Radiology Object in COntext version 2 (ROCOv2), a multimodal dataset consisting of radiological images and associated medical concepts and captions extracted from the PMC Open Access subset. It is an updated version of the ROCO dataset published in 2018, and adds 35,705 new images added to PMC since 2018. It further provides manually curated concepts for imaging modalities with additional anatomical and directional concepts for X-rays. The dataset consists of 79,789 images and has been used, with minor modifications, in the concept detection and caption prediction tasks of ImageCLEFmedical Caption 2023. The dataset is suitable for training image annotation models based on image-caption pairs, or for multi-label image classification using Unified Medical Language System (UMLS) concepts provided with each image. In addition, it can serve for pre-training of medical domain models, and evaluation of deep learning models for multi-task learning.
Comprehensive Study on German Language Models for Clinical and Biomedical Text Understanding
Apr 08, 2024



Abstract:Recent advances in natural language processing (NLP) can be largely attributed to the advent of pre-trained language models such as BERT and RoBERTa. While these models demonstrate remarkable performance on general datasets, they can struggle in specialized domains such as medicine, where unique domain-specific terminologies, domain-specific abbreviations, and varying document structures are common. This paper explores strategies for adapting these models to domain-specific requirements, primarily through continuous pre-training on domain-specific data. We pre-trained several German medical language models on 2.4B tokens derived from translated public English medical data and 3B tokens of German clinical data. The resulting models were evaluated on various German downstream tasks, including named entity recognition (NER), multi-label classification, and extractive question answering. Our results suggest that models augmented by clinical and translation-based pre-training typically outperform general domain models in medical contexts. We conclude that continuous pre-training has demonstrated the ability to match or even exceed the performance of clinical models trained from scratch. Furthermore, pre-training on clinical data or leveraging translated texts have proven to be reliable methods for domain adaptation in medical NLP tasks.
On the Impact of Cross-Domain Data on German Language Models
Oct 13, 2023Abstract:Traditionally, large language models have been either trained on general web crawls or domain-specific data. However, recent successes of generative large language models, have shed light on the benefits of cross-domain datasets. To examine the significance of prioritizing data diversity over quality, we present a German dataset comprising texts from five domains, along with another dataset aimed at containing high-quality data. Through training a series of models ranging between 122M and 750M parameters on both datasets, we conduct a comprehensive benchmark on multiple downstream tasks. Our findings demonstrate that the models trained on the cross-domain dataset outperform those trained on quality data alone, leading to improvements up to $4.45\%$ over the previous state-of-the-art. The models are available at https://huggingface.co/ikim-uk-essen
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge