Asma Ben Abacha
DermaVQA-DAS: Dermatology Assessment Schema (DAS) & Datasets for Closed-Ended Question Answering & Segmentation in Patient-Generated Dermatology Images
Dec 30, 2025Abstract:Recent advances in dermatological image analysis have been driven by large-scale annotated datasets; however, most existing benchmarks focus on dermatoscopic images and lack patient-authored queries and clinical context, limiting their applicability to patient-centered care. To address this gap, we introduce DermaVQA-DAS, an extension of the DermaVQA dataset that supports two complementary tasks: closed-ended question answering (QA) and dermatological lesion segmentation. Central to this work is the Dermatology Assessment Schema (DAS), a novel expert-developed framework that systematically captures clinically meaningful dermatological features in a structured and standardized form. DAS comprises 36 high-level and 27 fine-grained assessment questions, with multiple-choice options in English and Chinese. Leveraging DAS, we provide expert-annotated datasets for both closed QA and segmentation and benchmark state-of-the-art multimodal models. For segmentation, we evaluate multiple prompting strategies and show that prompt design impacts performance: the default prompt achieves the best results under Mean-of-Max and Mean-of-Mean evaluation aggregation schemes, while an augmented prompt incorporating both patient query title and content yields the highest performance under majority-vote-based microscore evaluation, achieving a Jaccard index of 0.395 and a Dice score of 0.566 with BiomedParse. For closed-ended QA, overall performance is strong across models, with average accuracies ranging from 0.729 to 0.798; o3 achieves the best overall accuracy (0.798), closely followed by GPT-4.1 (0.796), while Gemini-1.5-Pro shows competitive performance within the Gemini family (0.783). We publicly release DermaVQA-DAS, the DAS schema, and evaluation protocols to support and accelerate future research in patient-centered dermatological vision-language modeling (https://osf.io/72rp3).
Overview of the MEDIQA-OE 2025 Shared Task on Medical Order Extraction from Doctor-Patient Consultations
Oct 30, 2025Abstract:Clinical documentation increasingly uses automatic speech recognition and summarization, yet converting conversations into actionable medical orders for Electronic Health Records remains unexplored. A solution to this problem can significantly reduce the documentation burden of clinicians and directly impact downstream patient care. We introduce the MEDIQA-OE 2025 shared task, the first challenge on extracting medical orders from doctor-patient conversations. Six teams participated in the shared task and experimented with a broad range of approaches, and both closed- and open-weight large language models (LLMs). In this paper, we describe the MEDIQA-OE task, dataset, final leaderboard ranking, and participants' solutions.
MORQA: Benchmarking Evaluation Metrics for Medical Open-Ended Question Answering
Sep 15, 2025



Abstract:Evaluating natural language generation (NLG) systems in the medical domain presents unique challenges due to the critical demands for accuracy, relevance, and domain-specific expertise. Traditional automatic evaluation metrics, such as BLEU, ROUGE, and BERTScore, often fall short in distinguishing between high-quality outputs, especially given the open-ended nature of medical question answering (QA) tasks where multiple valid responses may exist. In this work, we introduce MORQA (Medical Open-Response QA), a new multilingual benchmark designed to assess the effectiveness of NLG evaluation metrics across three medical visual and text-based QA datasets in English and Chinese. Unlike prior resources, our datasets feature 2-4+ gold-standard answers authored by medical professionals, along with expert human ratings for three English and Chinese subsets. We benchmark both traditional metrics and large language model (LLM)-based evaluators, such as GPT-4 and Gemini, finding that LLM-based approaches significantly outperform traditional metrics in correlating with expert judgments. We further analyze factors driving this improvement, including LLMs' sensitivity to semantic nuances and robustness to variability among reference answers. Our results provide the first comprehensive, multilingual qualitative study of NLG evaluation in the medical domain, highlighting the need for human-aligned evaluation methods. All datasets and annotations will be publicly released to support future research.
A Modular Approach for Clinical SLMs Driven by Synthetic Data with Pre-Instruction Tuning, Model Merging, and Clinical-Tasks Alignment
May 15, 2025Abstract:High computation costs and latency of large language models such as GPT-4 have limited their deployment in clinical settings. Small language models (SLMs) offer a cost-effective alternative, but their limited capacity requires biomedical domain adaptation, which remains challenging. An additional bottleneck is the unavailability and high sensitivity of clinical data. To address these challenges, we propose a novel framework for adapting SLMs into high-performing clinical models. We introduce the MediPhi collection of 3.8B-parameter SLMs developed with our novel framework: pre-instruction tuning of experts on relevant medical and clinical corpora (PMC, Medical Guideline, MedWiki, etc.), model merging, and clinical-tasks alignment. To cover most clinical tasks, we extended the CLUE benchmark to CLUE+, doubling its size. Our expert models deliver relative improvements on this benchmark over the base model without any task-specific fine-tuning: 64.3% on medical entities, 49.5% on radiology reports, and 44% on ICD-10 coding (outperforming GPT-4-0125 by 14%). We unify the expert models into MediPhi via model merging, preserving gains across benchmarks. Furthermore, we built the MediFlow collection, a synthetic dataset of 2.5 million high-quality instructions on 14 medical NLP tasks, 98 fine-grained document types, and JSON format support. Alignment of MediPhi using supervised fine-tuning and direct preference optimization achieves further gains of 18.9% on average.
MEDEC: A Benchmark for Medical Error Detection and Correction in Clinical Notes
Dec 26, 2024Abstract:Several studies showed that Large Language Models (LLMs) can answer medical questions correctly, even outperforming the average human score in some medical exams. However, to our knowledge, no study has been conducted to assess the ability of language models to validate existing or generated medical text for correctness and consistency. In this paper, we introduce MEDEC (https://github.com/abachaa/MEDEC), the first publicly available benchmark for medical error detection and correction in clinical notes, covering five types of errors (Diagnosis, Management, Treatment, Pharmacotherapy, and Causal Organism). MEDEC consists of 3,848 clinical texts, including 488 clinical notes from three US hospital systems that were not previously seen by any LLM. The dataset has been used for the MEDIQA-CORR shared task to evaluate seventeen participating systems [Ben Abacha et al., 2024]. In this paper, we describe the data creation methods and we evaluate recent LLMs (e.g., o1-preview, GPT-4, Claude 3.5 Sonnet, and Gemini 2.0 Flash) for the tasks of detecting and correcting medical errors requiring both medical knowledge and reasoning capabilities. We also conducted a comparative study where two medical doctors performed the same task on the MEDEC test set. The results showed that MEDEC is a sufficiently challenging benchmark to assess the ability of models to validate existing or generated notes and to correct medical errors. We also found that although recent LLMs have a good performance in error detection and correction, they are still outperformed by medical doctors in these tasks. We discuss the potential factors behind this gap, the insights from our experiments, the limitations of current evaluation metrics, and share potential pointers for future research.
MedImageInsight: An Open-Source Embedding Model for General Domain Medical Imaging
Oct 09, 2024
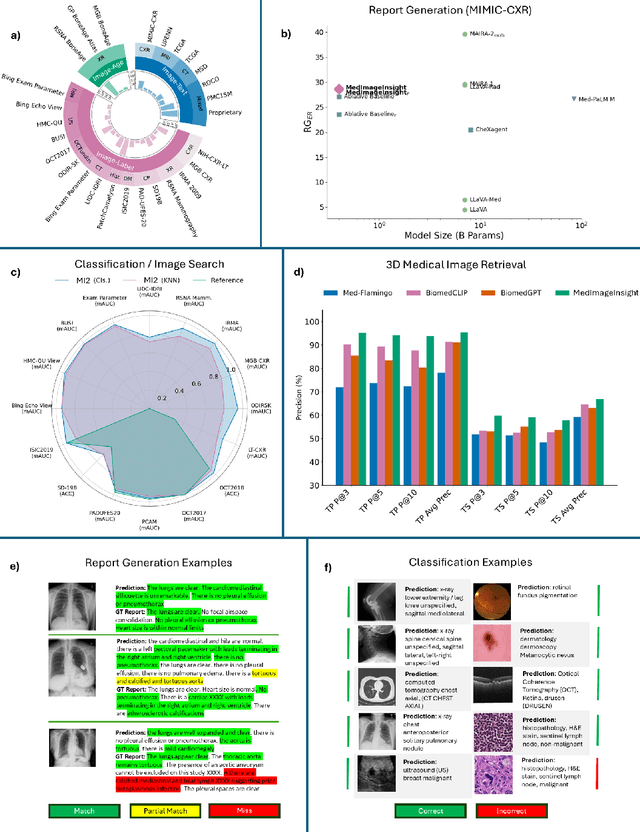
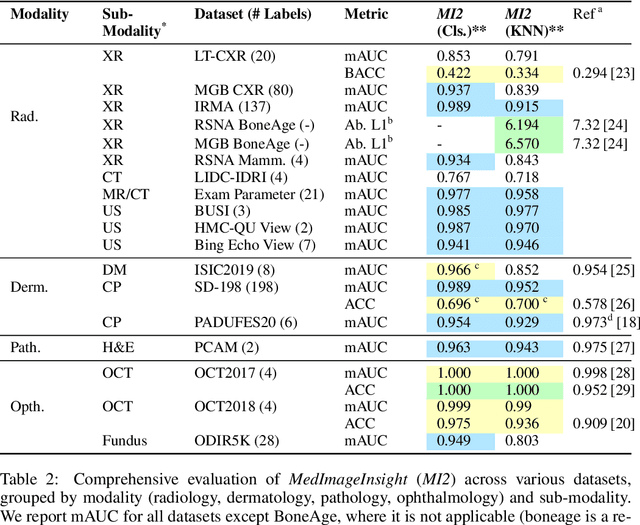
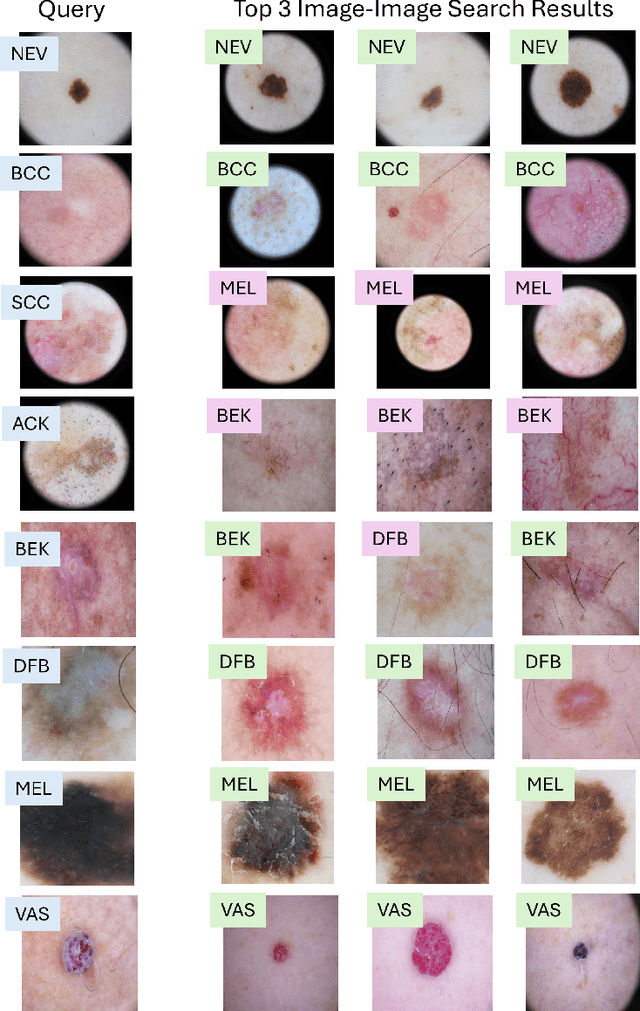
Abstract:In this work, we present MedImageInsight, an open-source medical imaging embedding model. MedImageInsight is trained on medical images with associated text and labels across a diverse collection of domains, including X-Ray, CT, MRI, dermoscopy, OCT, fundus photography, ultrasound, histopathology, and mammography. Rigorous evaluations demonstrate MedImageInsight's ability to achieve state-of-the-art (SOTA) or human expert level performance across classification, image-image search, and fine-tuning tasks. Specifically, on public datasets, MedImageInsight achieves SOTA in CT 3D medical image retrieval, as well as SOTA in disease classification and search for chest X-ray, dermatology, and OCT imaging. Furthermore, MedImageInsight achieves human expert performance in bone age estimation (on both public and partner data), as well as AUC above 0.9 in most other domains. When paired with a text decoder, MedImageInsight achieves near SOTA level single image report findings generation with less than 10\% the parameters of other models. Compared to fine-tuning GPT-4o with only MIMIC-CXR data for the same task, MedImageInsight outperforms in clinical metrics, but underperforms on lexical metrics where GPT-4o sets a new SOTA. Importantly for regulatory purposes, MedImageInsight can generate ROC curves, adjust sensitivity and specificity based on clinical need, and provide evidence-based decision support through image-image search (which can also enable retrieval augmented generation). In an independent clinical evaluation of image-image search in chest X-ray, MedImageInsight outperformed every other publicly available foundation model evaluated by large margins (over 6 points AUC), and significantly outperformed other models in terms of AI fairness (across age and gender). We hope releasing MedImageInsight will help enhance collective progress in medical imaging AI research and development.
ROCOv2: Radiology Objects in COntext Version 2, an Updated Multimodal Image Dataset
May 16, 2024Abstract:Automated medical image analysis systems often require large amounts of training data with high quality labels, which are difficult and time consuming to generate. This paper introduces Radiology Object in COntext version 2 (ROCOv2), a multimodal dataset consisting of radiological images and associated medical concepts and captions extracted from the PMC Open Access subset. It is an updated version of the ROCO dataset published in 2018, and adds 35,705 new images added to PMC since 2018. It further provides manually curated concepts for imaging modalities with additional anatomical and directional concepts for X-rays. The dataset consists of 79,789 images and has been used, with minor modifications, in the concept detection and caption prediction tasks of ImageCLEFmedical Caption 2023. The dataset is suitable for training image annotation models based on image-caption pairs, or for multi-label image classification using Unified Medical Language System (UMLS) concepts provided with each image. In addition, it can serve for pre-training of medical domain models, and evaluation of deep learning models for multi-task learning.
3D-MIR: A Benchmark and Empirical Study on 3D Medical Image Retrieval in Radiology
Nov 23, 2023



Abstract:The increasing use of medical imaging in healthcare settings presents a significant challenge due to the increasing workload for radiologists, yet it also offers opportunity for enhancing healthcare outcomes if effectively leveraged. 3D image retrieval holds potential to reduce radiologist workloads by enabling clinicians to efficiently search through diagnostically similar or otherwise relevant cases, resulting in faster and more precise diagnoses. However, the field of 3D medical image retrieval is still emerging, lacking established evaluation benchmarks, comprehensive datasets, and thorough studies. This paper attempts to bridge this gap by introducing a novel benchmark for 3D Medical Image Retrieval (3D-MIR) that encompasses four different anatomies imaged with computed tomography. Using this benchmark, we explore a diverse set of search strategies that use aggregated 2D slices, 3D volumes, and multi-modal embeddings from popular multi-modal foundation models as queries. Quantitative and qualitative assessments of each approach are provided alongside an in-depth discussion that offers insight for future research. To promote the advancement of this field, our benchmark, dataset, and code are made publicly available.
ACI-BENCH: a Novel Ambient Clinical Intelligence Dataset for Benchmarking Automatic Visit Note Generation
Jun 03, 2023Abstract:Recent immense breakthroughs in generative models such as in GPT4 have precipitated re-imagined ubiquitous usage of these models in all applications. One area that can benefit by improvements in artificial intelligence (AI) is healthcare. The note generation task from doctor-patient encounters, and its associated electronic medical record documentation, is one of the most arduous time-consuming tasks for physicians. It is also a natural prime potential beneficiary to advances in generative models. However with such advances, benchmarking is more critical than ever. Whether studying model weaknesses or developing new evaluation metrics, shared open datasets are an imperative part of understanding the current state-of-the-art. Unfortunately as clinic encounter conversations are not routinely recorded and are difficult to ethically share due to patient confidentiality, there are no sufficiently large clinic dialogue-note datasets to benchmark this task. Here we present the Ambient Clinical Intelligence Benchmark (ACI-BENCH) corpus, the largest dataset to date tackling the problem of AI-assisted note generation from visit dialogue. We also present the benchmark performances of several common state-of-the-art approaches.
An Investigation of Evaluation Metrics for Automated Medical Note Generation
May 27, 2023



Abstract:Recent studies on automatic note generation have shown that doctors can save significant amounts of time when using automatic clinical note generation (Knoll et al., 2022). Summarization models have been used for this task to generate clinical notes as summaries of doctor-patient conversations (Krishna et al., 2021; Cai et al., 2022). However, assessing which model would best serve clinicians in their daily practice is still a challenging task due to the large set of possible correct summaries, and the potential limitations of automatic evaluation metrics. In this paper, we study evaluation methods and metrics for the automatic generation of clinical notes from medical conversations. In particular, we propose new task-specific metrics and we compare them to SOTA evaluation metrics in text summarization and generation, including: (i) knowledge-graph embedding-based metrics, (ii) customized model-based metrics, (iii) domain-adapted/fine-tuned metrics, and (iv) ensemble metrics. To study the correlation between the automatic metrics and manual judgments, we evaluate automatic notes/summaries by comparing the system and reference facts and computing the factual correctness, and the hallucination and omission rates for critical medical facts. This study relied on seven datasets manually annotated by domain experts. Our experiments show that automatic evaluation metrics can have substantially different behaviors on different types of clinical notes datasets. However, the results highlight one stable subset of metrics as the most correlated with human judgments with a relevant aggregation of different evaluation criteria.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge