George Michalopoulos
An Investigation of Evaluation Metrics for Automated Medical Note Generation
May 27, 2023



Abstract:Recent studies on automatic note generation have shown that doctors can save significant amounts of time when using automatic clinical note generation (Knoll et al., 2022). Summarization models have been used for this task to generate clinical notes as summaries of doctor-patient conversations (Krishna et al., 2021; Cai et al., 2022). However, assessing which model would best serve clinicians in their daily practice is still a challenging task due to the large set of possible correct summaries, and the potential limitations of automatic evaluation metrics. In this paper, we study evaluation methods and metrics for the automatic generation of clinical notes from medical conversations. In particular, we propose new task-specific metrics and we compare them to SOTA evaluation metrics in text summarization and generation, including: (i) knowledge-graph embedding-based metrics, (ii) customized model-based metrics, (iii) domain-adapted/fine-tuned metrics, and (iv) ensemble metrics. To study the correlation between the automatic metrics and manual judgments, we evaluate automatic notes/summaries by comparing the system and reference facts and computing the factual correctness, and the hallucination and omission rates for critical medical facts. This study relied on seven datasets manually annotated by domain experts. Our experiments show that automatic evaluation metrics can have substantially different behaviors on different types of clinical notes datasets. However, the results highlight one stable subset of metrics as the most correlated with human judgments with a relevant aggregation of different evaluation criteria.
ICDBigBird: A Contextual Embedding Model for ICD Code Classification
Apr 21, 2022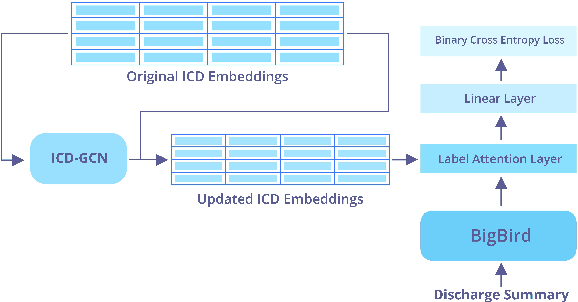
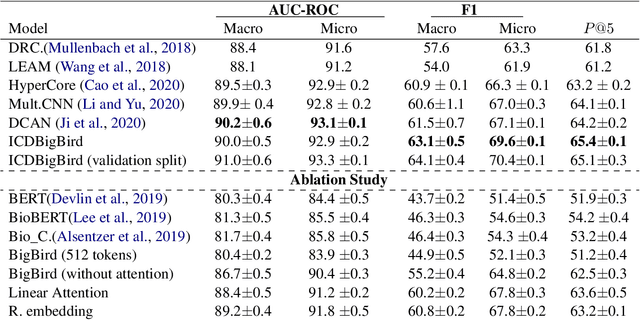
Abstract:The International Classification of Diseases (ICD) system is the international standard for classifying diseases and procedures during a healthcare encounter and is widely used for healthcare reporting and management purposes. Assigning correct codes for clinical procedures is important for clinical, operational, and financial decision-making in healthcare. Contextual word embedding models have achieved state-of-the-art results in multiple NLP tasks. However, these models have yet to achieve state-of-the-art results in the ICD classification task since one of their main disadvantages is that they can only process documents that contain a small number of tokens which is rarely the case with real patient notes. In this paper, we introduce ICDBigBird a BigBird-based model which can integrate a Graph Convolutional Network (GCN), that takes advantage of the relations between ICD codes in order to create 'enriched' representations of their embeddings, with a BigBird contextual model that can process larger documents. Our experiments on a real-world clinical dataset demonstrate the effectiveness of our BigBird-based model on the ICD classification task as it outperforms the previous state-of-the-art models.
Cohort Characteristics and Factors Associated with Cannabis Use among Adolescents in Canada Using Pattern Discovery and Disentanglement Method
Sep 03, 2021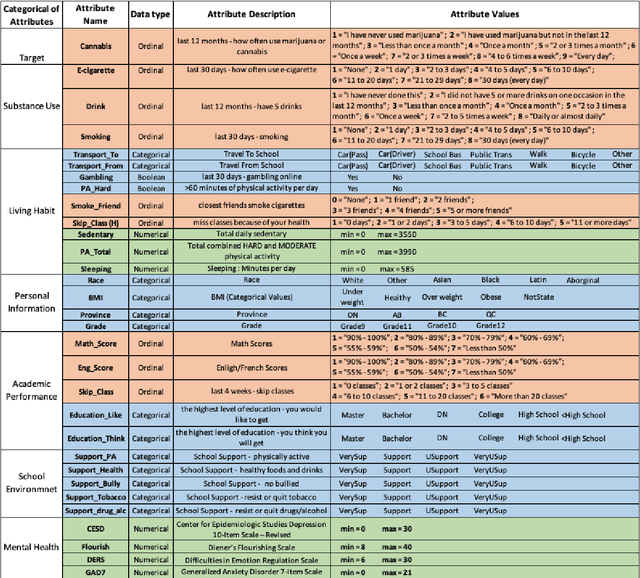
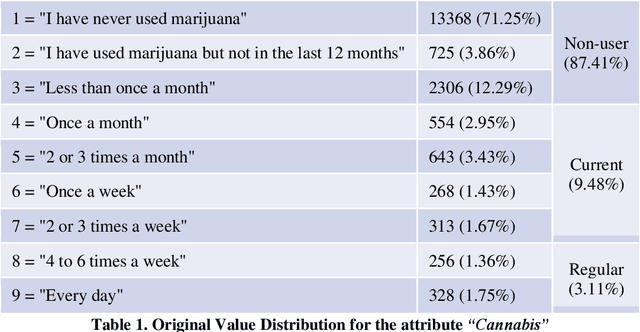

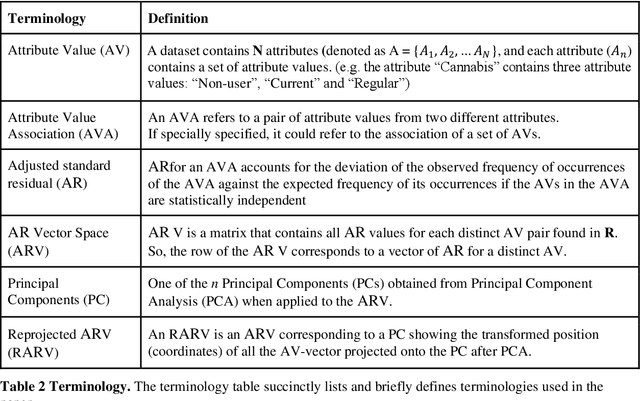
Abstract:COMPASS is a longitudinal, prospective cohort study collecting data annually from students attending high school in jurisdictions across Canada. We aimed to discover significant frequent/rare associations of behavioral factors among Canadian adolescents related to cannabis use. We use a subset of COMPASS dataset which contains 18,761 records of students in grades 9 to 12 with 31 selected features (attributes) involving various characteristics, from living habits to academic performance. We then used the Pattern Discovery and Disentanglement (PDD) algorithm that we have developed to detect strong and rare (yet statistically significant) associations from the dataset. PDD used the criteria derived from disentangled statistical spaces (known as Re-projected Adjusted-Standardized Residual Vector Spaces, notated as RARV). It outperformed methods using other criteria (i.e. support and confidence) popular as reported in the literature. Association results showed that PDD can discover: i) a smaller set of succinct significant associations in clusters; ii) frequent and rare, yet significant, patterns supported by population health relevant study; iii) patterns from a dataset with extremely imbalanced groups (majority class: minority class = 88.3%: 11.7%).
LexSubCon: Integrating Knowledge from Lexical Resources into Contextual Embeddings for Lexical Substitution
Jul 11, 2021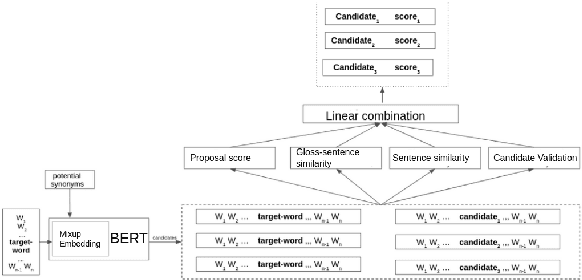
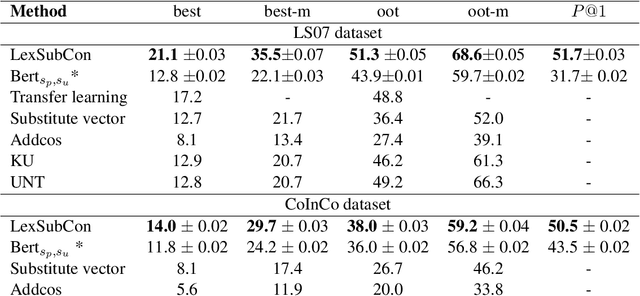
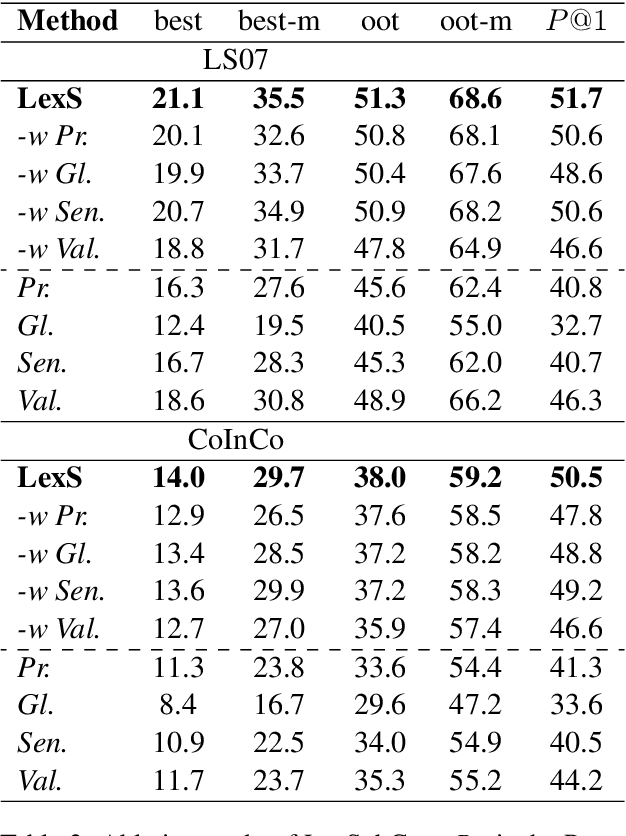
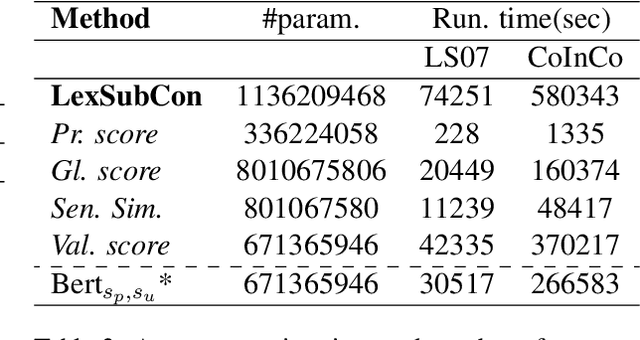
Abstract:Lexical substitution is the task of generating meaningful substitutes for a word in a given textual context. Contextual word embedding models have achieved state-of-the-art results in the lexical substitution task by relying on contextual information extracted from the replaced word within the sentence. However, such models do not take into account structured knowledge that exists in external lexical databases. We introduce LexSubCon, an end-to-end lexical substitution framework based on contextual embedding models that can identify highly accurate substitute candidates. This is achieved by combining contextual information with knowledge from structured lexical resources. Our approach involves: (i) introducing a novel mix-up embedding strategy in the creation of the input embedding of the target word through linearly interpolating the pair of the target input embedding and the average embedding of its probable synonyms; (ii) considering the similarity of the sentence-definition embeddings of the target word and its proposed candidates; and, (iii) calculating the effect of each substitution in the semantics of the sentence through a fine-tuned sentence similarity model. Our experiments show that LexSubCon outperforms previous state-of-the-art methods on LS07 and CoInCo benchmark datasets that are widely used for lexical substitution tasks.
UmlsBERT: Clinical Domain Knowledge Augmentation of Contextual Embeddings Using the Unified Medical Language System Metathesaurus
Oct 20, 2020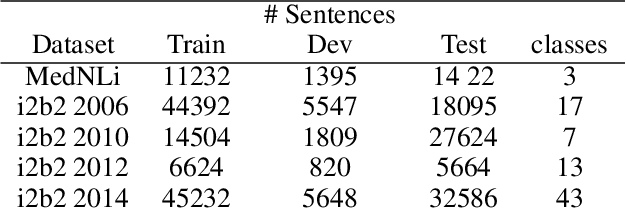
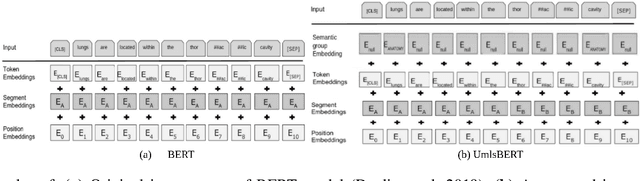
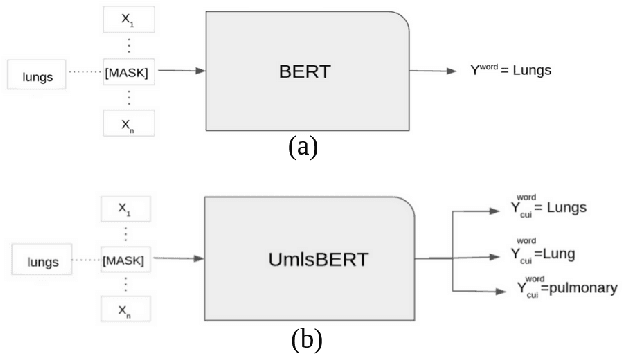
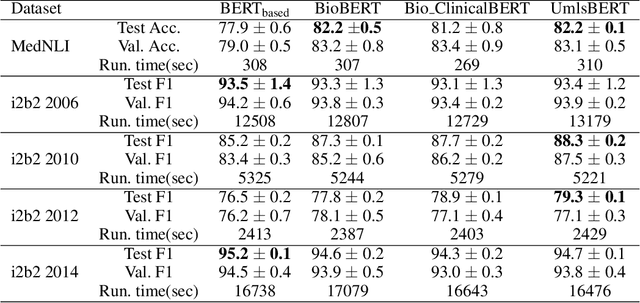
Abstract:Contextual word embedding models, such as BioBERT and Bio_ClinicalBERT, have achieved state-of-the-art results in biomedical natural language processing tasks by focusing their pre-training process on domain-specific corpora. However, such models do not take into consideration expert domain knowledge. In this work, we introduced UmlsBERT, a contextual embedding model that integrates domain knowledge during the pre-training process via a novel knowledge augmentation strategy. More specifically, the augmentation on UmlsBERT with the Unified Medical Language System (UMLS) Metathesaurus was performed in two ways: i) connecting words that have the same underlying `concept' in UMLS, and ii) leveraging semantic group knowledge in UMLS to create clinically meaningful input embeddings. By applying these two strategies, UmlsBERT can encode clinical domain knowledge into word embeddings and outperform existing domain-specific models on common named-entity recognition (NER) and clinical natural language inference clinical NLP tasks.
Where's the Question? A Multi-channel Deep Convolutional Neural Network for Question Identification in Textual Data
Oct 15, 2020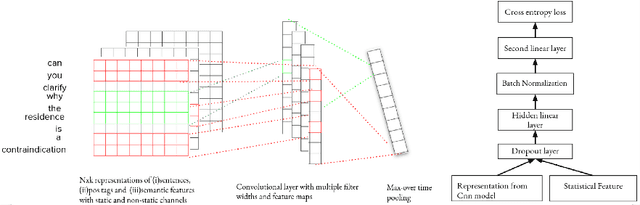
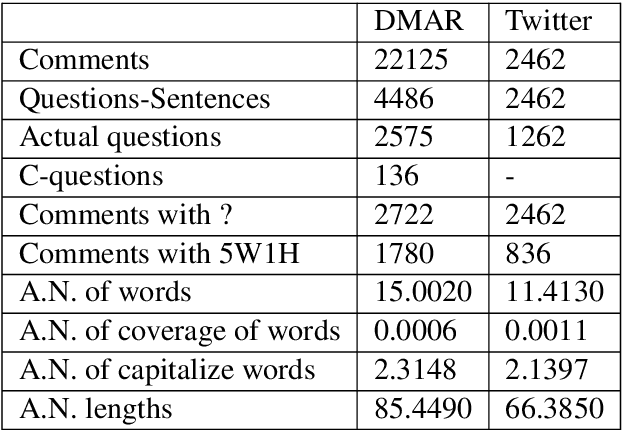
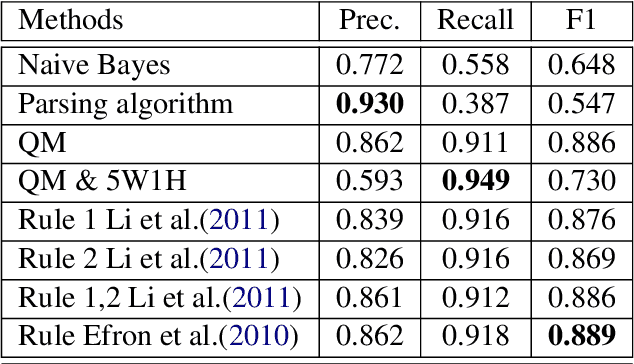
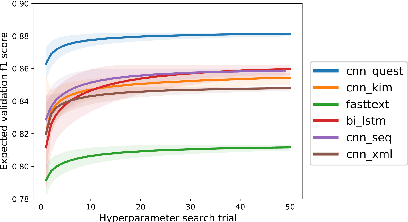
Abstract:In most clinical practice settings, there is no rigorous reviewing of the clinical documentation, resulting in inaccurate information captured in the patient medical records. The gold standard in clinical data capturing is achieved via "expert-review", where clinicians can have a dialogue with a domain expert (reviewers) and ask them questions about data entry rules. Automatically identifying "real questions" in these dialogues could uncover ambiguities or common problems in data capturing in a given clinical setting. In this study, we proposed a novel multi-channel deep convolutional neural network architecture, namely Quest-CNN, for the purpose of separating real questions that expect an answer (information or help) about an issue from sentences that are not questions, as well as from questions referring to an issue mentioned in a nearby sentence (e.g., can you clarify this?), which we will refer as "c-questions". We conducted a comprehensive performance comparison analysis of the proposed multi-channel deep convolutional neural network against other deep neural networks. Furthermore, we evaluated the performance of traditional rule-based and learning-based methods for detecting question sentences. The proposed Quest-CNN achieved the best F1 score both on a dataset of data entry-review dialogue in a dialysis care setting, and on a general domain dataset.
Record fusion: A learning approach
Jun 18, 2020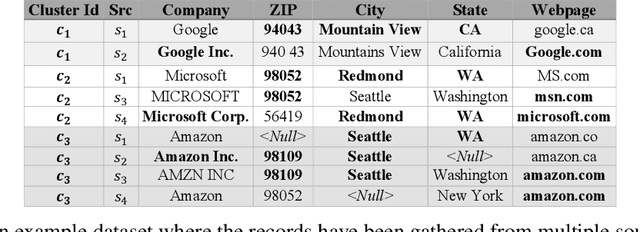
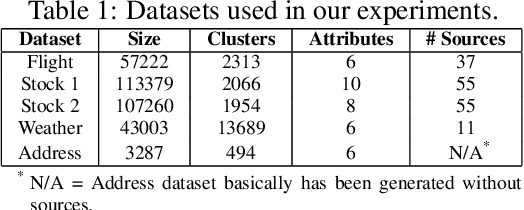
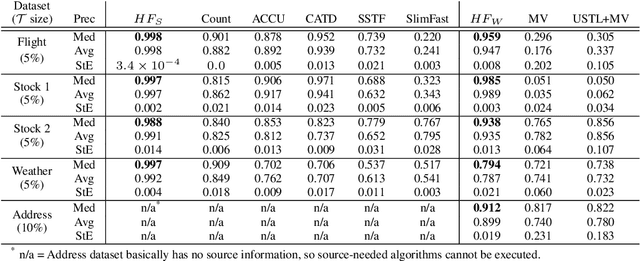
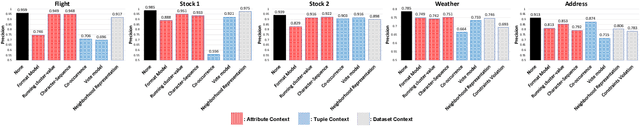
Abstract:Record fusion is the task of aggregating multiple records that correspond to the same real-world entity in a database. We can view record fusion as a machine learning problem where the goal is to predict the "correct" value for each attribute for each entity. Given a database, we use a combination of attribute-level, recordlevel, and database-level signals to construct a feature vector for each cell (or (row, col)) of that database. We use this feature vector alongwith the ground-truth information to learn a classifier for each of the attributes of the database. Our learning algorithm uses a novel stagewise additive model. At each stage, we construct a new feature vector by combining a part of the original feature vector with features computed by the predictions from the previous stage. We then learn a softmax classifier over the new feature space. This greedy stagewise approach can be viewed as a deep model where at each stage, we are adding more complicated non-linear transformations of the original feature vector. We show that our approach fuses records with an average precision of ~98% when source information of records is available, and ~94% without source information across a diverse array of real-world datasets. We compare our approach to a comprehensive collection of data fusion and entity consolidation methods considered in the literature. We show that our approach can achieve an average precision improvement of ~20%/~45% with/without source information respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge