Nicola Sahar
ICDBigBird: A Contextual Embedding Model for ICD Code Classification
Apr 21, 2022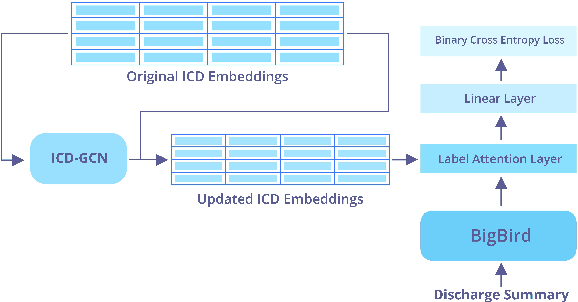
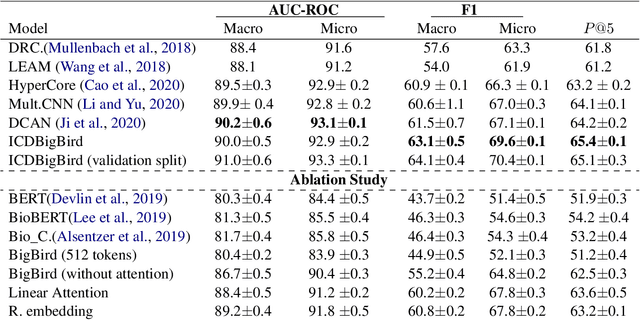
Abstract:The International Classification of Diseases (ICD) system is the international standard for classifying diseases and procedures during a healthcare encounter and is widely used for healthcare reporting and management purposes. Assigning correct codes for clinical procedures is important for clinical, operational, and financial decision-making in healthcare. Contextual word embedding models have achieved state-of-the-art results in multiple NLP tasks. However, these models have yet to achieve state-of-the-art results in the ICD classification task since one of their main disadvantages is that they can only process documents that contain a small number of tokens which is rarely the case with real patient notes. In this paper, we introduce ICDBigBird a BigBird-based model which can integrate a Graph Convolutional Network (GCN), that takes advantage of the relations between ICD codes in order to create 'enriched' representations of their embeddings, with a BigBird contextual model that can process larger documents. Our experiments on a real-world clinical dataset demonstrate the effectiveness of our BigBird-based model on the ICD classification task as it outperforms the previous state-of-the-art models.
Active learning for medical code assignment
Apr 12, 2021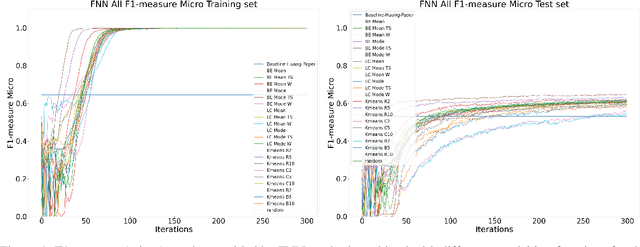
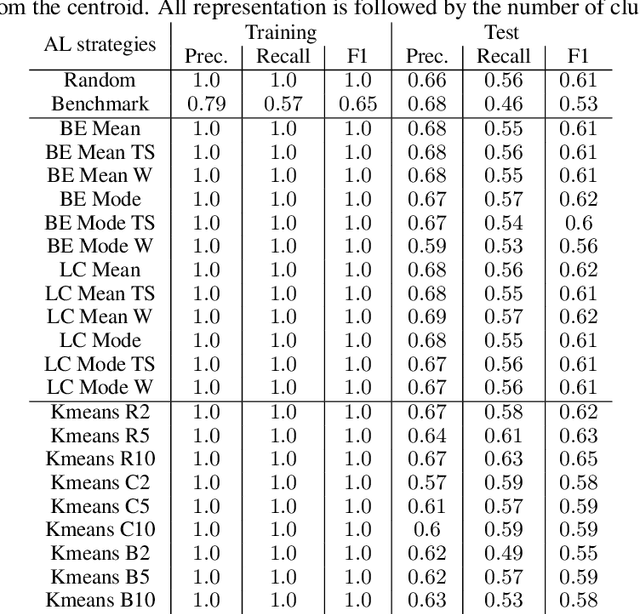
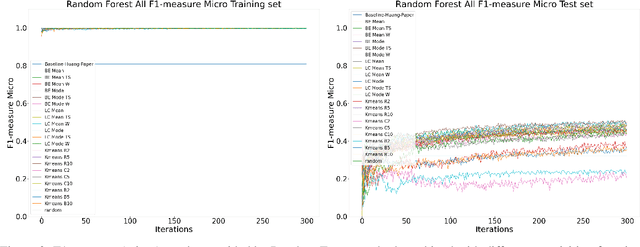
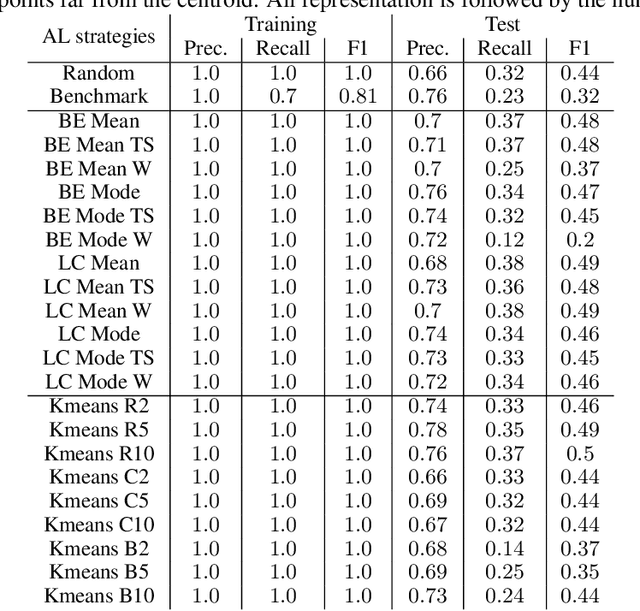
Abstract:Machine Learning (ML) is widely used to automatically extract meaningful information from Electronic Health Records (EHR) to support operational, clinical, and financial decision-making. However, ML models require a large number of annotated examples to provide satisfactory results, which is not possible in most healthcare scenarios due to the high cost of clinician-labeled data. Active Learning (AL) is a process of selecting the most informative instances to be labeled by an expert to further train a supervised algorithm. We demonstrate the effectiveness of AL in multi-label text classification in the clinical domain. In this context, we apply a set of well-known AL methods to help automatically assign ICD-9 codes on the MIMIC-III dataset. Our results show that the selection of informative instances provides satisfactory classification with a significantly reduced training set (8.3\% of the total instances). We conclude that AL methods can significantly reduce the manual annotation cost while preserving model performance.
End-to-end Named Entity Recognition and Relation Extraction using Pre-trained Language Models
Dec 20, 2019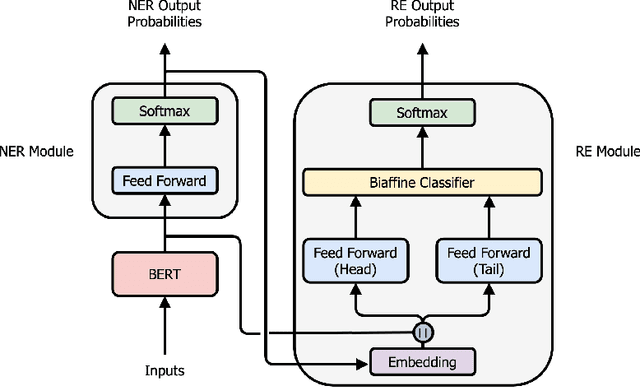
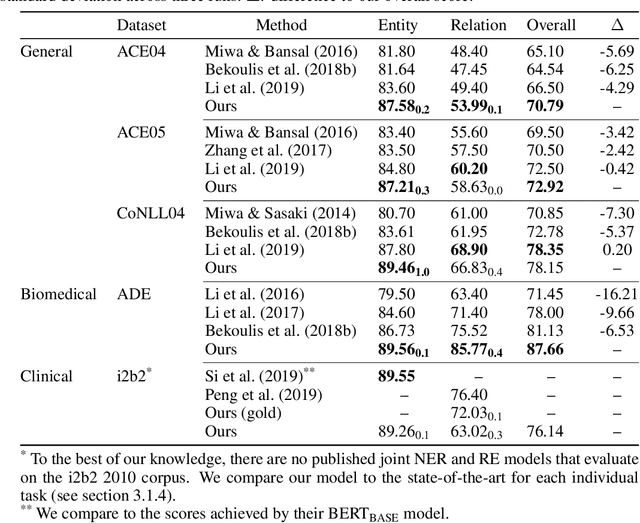
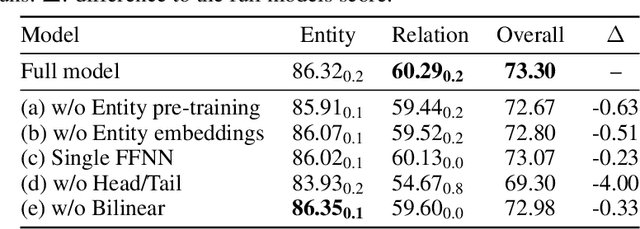
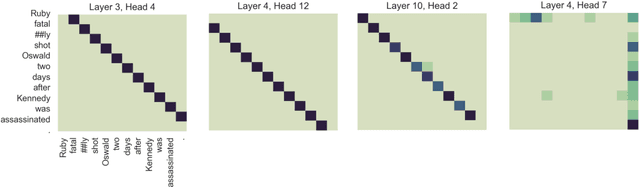
Abstract:Named entity recognition (NER) and relation extraction (RE) are two important tasks in information extraction and retrieval (IE \& IR). Recent work has demonstrated that it is beneficial to learn these tasks jointly, which avoids the propagation of error inherent in pipeline-based systems and improves performance. However, state-of-the-art joint models typically rely on external natural language processing (NLP) tools, such as dependency parsers, limiting their usefulness to domains (e.g. news) where those tools perform well. The few neural, end-to-end models that have been proposed are trained almost completely from scratch. In this paper, we propose a neural, end-to-end model for jointly extracting entities and their relations which does not rely on external NLP tools and which integrates a large, pre-trained language model. Because the bulk of our model's parameters are pre-trained and we eschew recurrence for self-attention, our model is fast to train. On 5 datasets across 3 domains, our model matches or exceeds state-of-the-art performance, sometimes by a large margin.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge