Xindi Wang
A Domain-Adapted Pipeline for Structured Information Extraction from Police Incident Announcements on Social Media
Dec 18, 2025Abstract:Structured information extraction from police incident announcements is crucial for timely and accurate data processing, yet presents considerable challenges due to the variability and informal nature of textual sources such as social media posts. To address these challenges, we developed a domain-adapted extraction pipeline that leverages targeted prompt engineering with parameter-efficient fine-tuning of the Qwen2.5-7B model using Low-Rank Adaptation (LoRA). This approach enables the model to handle noisy, heterogeneous text while reliably extracting 15 key fields, including location, event characteristics, and impact assessment, from a high-quality, manually annotated dataset of 4,933 instances derived from 27,822 police briefing posts on Chinese Weibo (2019-2020). Experimental results demonstrated that LoRA-based fine-tuning significantly improved performance over both the base and instruction-tuned models, achieving an accuracy exceeding 98.36% for mortality detection and Exact Match Rates of 95.31% for fatality counts and 95.54% for province-level location extraction. The proposed pipeline thus provides a validated and efficient solution for multi-task structured information extraction in specialized domains, offering a practical framework for transforming unstructured text into reliable structured data in social science research.
ChineseEEG-2: An EEG Dataset for Multimodal Semantic Alignment and Neural Decoding during Reading and Listening
Aug 06, 2025Abstract:EEG-based neural decoding requires large-scale benchmark datasets. Paired brain-language data across speaking, listening, and reading modalities are essential for aligning neural activity with the semantic representation of large language models (LLMs). However, such datasets are rare, especially for non-English languages. Here, we present ChineseEEG-2, a high-density EEG dataset designed for benchmarking neural decoding models under real-world language tasks. Building on our previous ChineseEEG dataset, which focused on silent reading, ChineseEEG-2 adds two active modalities: Reading Aloud (RA) and Passive Listening (PL), using the same Chinese corpus. EEG and audio were simultaneously recorded from four participants during ~10.7 hours of reading aloud. These recordings were then played to eight other participants, collecting ~21.6 hours of EEG during listening. This setup enables speech temporal and semantic alignment across the RA and PL modalities. ChineseEEG-2 includes EEG signals, precise audio, aligned semantic embeddings from pre-trained language models, and task labels. Together with ChineseEEG, this dataset supports joint semantic alignment learning across speaking, listening, and reading. It enables benchmarking of neural decoding algorithms and promotes brain-LLM alignment under multimodal language tasks, especially in Chinese. ChineseEEG-2 provides a benchmark dataset for next-generation neural semantic decoding.
Generative Large Language Models Trained for Detecting Errors in Radiology Reports
Apr 06, 2025Abstract:In this retrospective study, a dataset was constructed with two parts. The first part included 1,656 synthetic chest radiology reports generated by GPT-4 using specified prompts, with 828 being error-free synthetic reports and 828 containing errors. The second part included 614 reports: 307 error-free reports between 2011 and 2016 from the MIMIC-CXR database and 307 corresponding synthetic reports with errors generated by GPT-4 on the basis of these MIMIC-CXR reports and specified prompts. All errors were categorized into four types: negation, left/right, interval change, and transcription errors. Then, several models, including Llama-3, GPT-4, and BiomedBERT, were refined using zero-shot prompting, few-shot prompting, or fine-tuning strategies. Finally, the performance of these models was evaluated using the F1 score, 95\% confidence interval (CI) and paired-sample t-tests on our constructed dataset, with the prediction results further assessed by radiologists. Using zero-shot prompting, the fine-tuned Llama-3-70B-Instruct model achieved the best performance with the following F1 scores: 0.769 for negation errors, 0.772 for left/right errors, 0.750 for interval change errors, 0.828 for transcription errors, and 0.780 overall. In the real-world evaluation phase, two radiologists reviewed 200 randomly selected reports output by the model. Of these, 99 were confirmed to contain errors detected by the models by both radiologists, and 163 were confirmed to contain model-detected errors by at least one radiologist. Generative LLMs, fine-tuned on synthetic and MIMIC-CXR radiology reports, greatly enhanced error detection in radiology reports.
Graph-tree Fusion Model with Bidirectional Information Propagation for Long Document Classification
Oct 03, 2024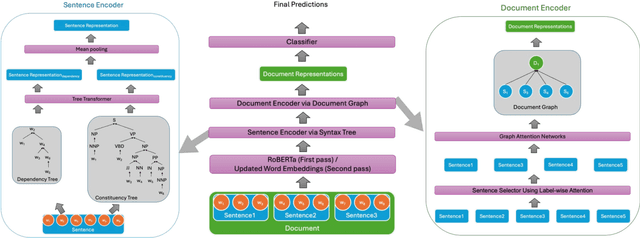
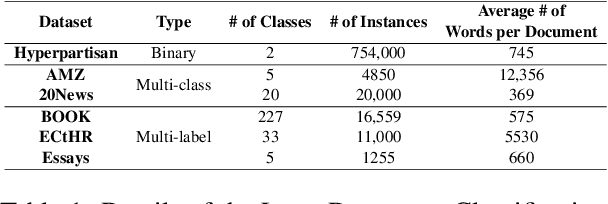
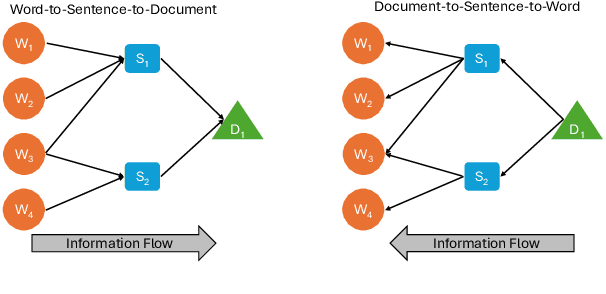
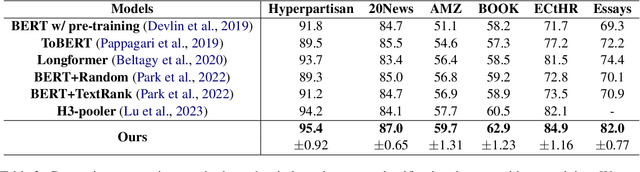
Abstract:Long document classification presents challenges in capturing both local and global dependencies due to their extensive content and complex structure. Existing methods often struggle with token limits and fail to adequately model hierarchical relationships within documents. To address these constraints, we propose a novel model leveraging a graph-tree structure. Our approach integrates syntax trees for sentence encodings and document graphs for document encodings, which capture fine-grained syntactic relationships and broader document contexts, respectively. We use Tree Transformers to generate sentence encodings, while a graph attention network models inter- and intra-sentence dependencies. During training, we implement bidirectional information propagation from word-to-sentence-to-document and vice versa, which enriches the contextual representation. Our proposed method enables a comprehensive understanding of content at all hierarchical levels and effectively handles arbitrarily long contexts without token limit constraints. Experimental results demonstrate the effectiveness of our approach in all types of long document classification tasks.
Enhancing disease detection in radiology reports through fine-tuning lightweight LLM on weak labels
Sep 25, 2024Abstract:Despite significant progress in applying large language models (LLMs) to the medical domain, several limitations still prevent them from practical applications. Among these are the constraints on model size and the lack of cohort-specific labeled datasets. In this work, we investigated the potential of improving a lightweight LLM, such as Llama 3.1-8B, through fine-tuning with datasets using synthetic labels. Two tasks are jointly trained by combining their respective instruction datasets. When the quality of the task-specific synthetic labels is relatively high (e.g., generated by GPT4- o), Llama 3.1-8B achieves satisfactory performance on the open-ended disease detection task, with a micro F1 score of 0.91. Conversely, when the quality of the task-relevant synthetic labels is relatively low (e.g., from the MIMIC-CXR dataset), fine-tuned Llama 3.1-8B is able to surpass its noisy teacher labels (micro F1 score of 0.67 v.s. 0.63) when calibrated against curated labels, indicating the strong inherent underlying capability of the model. These findings demonstrate the potential of fine-tuning LLMs with synthetic labels, offering a promising direction for future research on LLM specialization in the medical domain.
Auxiliary Knowledge-Induced Learning for Automatic Multi-Label Medical Document Classification
May 29, 2024Abstract:The International Classification of Diseases (ICD) is an authoritative medical classification system of different diseases and conditions for clinical and management purposes. ICD indexing assigns a subset of ICD codes to a medical record. Since human coding is labour-intensive and error-prone, many studies employ machine learning to automate the coding process. ICD coding is a challenging task, as it needs to assign multiple codes to each medical document from an extremely large hierarchically organized collection. In this paper, we propose a novel approach for ICD indexing that adopts three ideas: (1) we use a multi-level deep dilated residual convolution encoder to aggregate the information from the clinical notes and learn document representations across different lengths of the texts; (2) we formalize the task of ICD classification with auxiliary knowledge of the medical records, which incorporates not only the clinical texts but also different clinical code terminologies and drug prescriptions for better inferring the ICD codes; and (3) we introduce a graph convolutional network to leverage the co-occurrence patterns among ICD codes, aiming to enhance the quality of label representations. Experimental results show the proposed method achieves state-of-the-art performance on a number of measures.
Multi-stage Retrieve and Re-rank Model for Automatic Medical Coding Recommendation
May 29, 2024
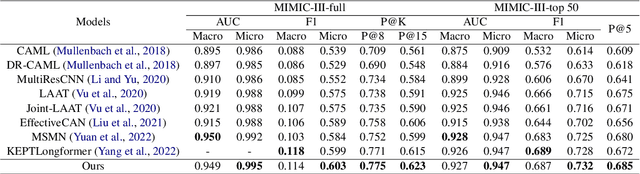
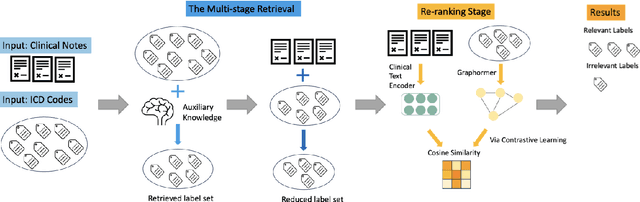

Abstract:The International Classification of Diseases (ICD) serves as a definitive medical classification system encompassing a wide range of diseases and conditions. The primary objective of ICD indexing is to allocate a subset of ICD codes to a medical record, which facilitates standardized documentation and management of various health conditions. Most existing approaches have suffered from selecting the proper label subsets from an extremely large ICD collection with a heavy long-tailed label distribution. In this paper, we leverage a multi-stage ``retrieve and re-rank'' framework as a novel solution to ICD indexing, via a hybrid discrete retrieval method, and re-rank retrieved candidates with contrastive learning that allows the model to make more accurate predictions from a simplified label space. The retrieval model is a hybrid of auxiliary knowledge of the electronic health records (EHR) and a discrete retrieval method (BM25), which efficiently collects high-quality candidates. In the last stage, we propose a label co-occurrence guided contrastive re-ranking model, which re-ranks the candidate labels by pulling together the clinical notes with positive ICD codes. Experimental results show the proposed method achieves state-of-the-art performance on a number of measures on the MIMIC-III benchmark.
Beyond the Limits: A Survey of Techniques to Extend the Context Length in Large Language Models
Feb 03, 2024Abstract:Recently, large language models (LLMs) have shown remarkable capabilities including understanding context, engaging in logical reasoning, and generating responses. However, this is achieved at the expense of stringent computational and memory requirements, hindering their ability to effectively support long input sequences. This survey provides an inclusive review of the recent techniques and methods devised to extend the sequence length in LLMs, thereby enhancing their capacity for long-context understanding. In particular, we review and categorize a wide range of techniques including architectural modifications, such as modified positional encoding and altered attention mechanisms, which are designed to enhance the processing of longer sequences while avoiding a proportional increase in computational requirements. The diverse methodologies investigated in this study can be leveraged across different phases of LLMs, i.e., training, fine-tuning and inference. This enables LLMs to efficiently process extended sequences. The limitations of the current methodologies is discussed in the last section along with the suggestions for future research directions, underscoring the importance of sequence length in the continued advancement of LLMs.
Investigating the Learning Behaviour of In-context Learning: A Comparison with Supervised Learning
Aug 01, 2023Abstract:Large language models (LLMs) have shown remarkable capacity for in-context learning (ICL), where learning a new task from just a few training examples is done without being explicitly pre-trained. However, despite the success of LLMs, there has been little understanding of how ICL learns the knowledge from the given prompts. In this paper, to make progress toward understanding the learning behaviour of ICL, we train the same LLMs with the same demonstration examples via ICL and supervised learning (SL), respectively, and investigate their performance under label perturbations (i.e., noisy labels and label imbalance) on a range of classification tasks. First, via extensive experiments, we find that gold labels have significant impacts on the downstream in-context performance, especially for large language models; however, imbalanced labels matter little to ICL across all model sizes. Second, when comparing with SL, we show empirically that ICL is less sensitive to label perturbations than SL, and ICL gradually attains comparable performance to SL as the model size increases.
MeSHup: A Corpus for Full Text Biomedical Document Indexing
Apr 28, 2022
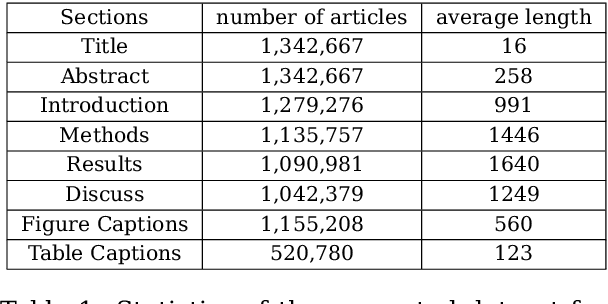
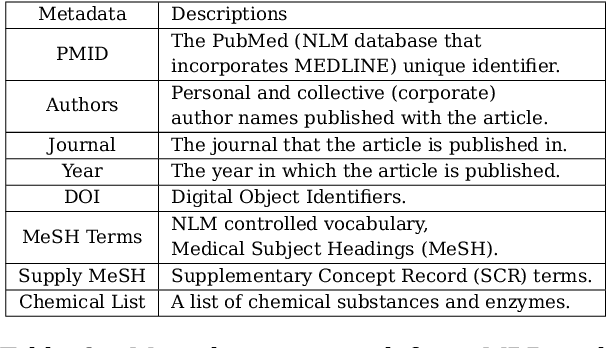
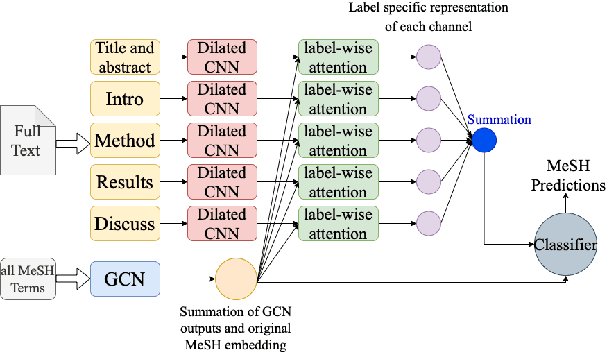
Abstract:Medical Subject Heading (MeSH) indexing refers to the problem of assigning a given biomedical document with the most relevant labels from an extremely large set of MeSH terms. Currently, the vast number of biomedical articles in the PubMed database are manually annotated by human curators, which is time consuming and costly; therefore, a computational system that can assist the indexing is highly valuable. When developing supervised MeSH indexing systems, the availability of a large-scale annotated text corpus is desirable. A publicly available, large corpus that permits robust evaluation and comparison of various systems is important to the research community. We release a large scale annotated MeSH indexing corpus, MeSHup, which contains 1,342,667 full text articles in English, together with the associated MeSH labels and metadata, authors, and publication venues that are collected from the MEDLINE database. We train an end-to-end model that combines features from documents and their associated labels on our corpus and report the new baseline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge