KenMeSH: Knowledge-enhanced End-to-end Biomedical Text Labelling
Paper and Code
Mar 14, 2022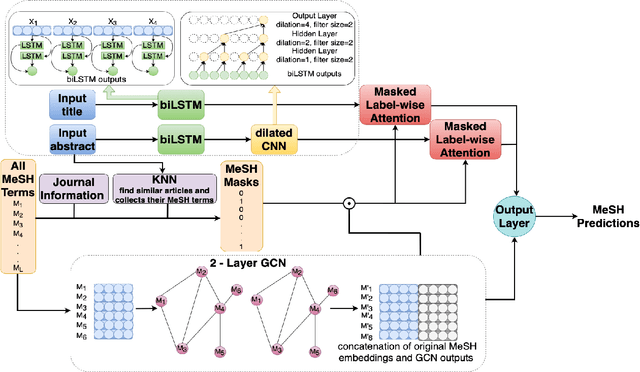
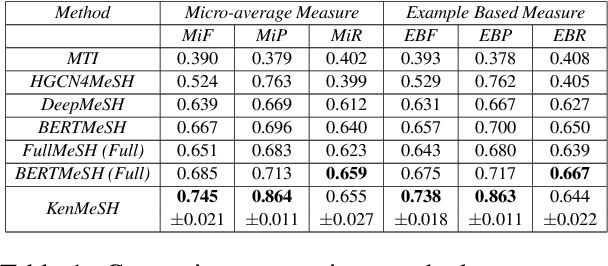
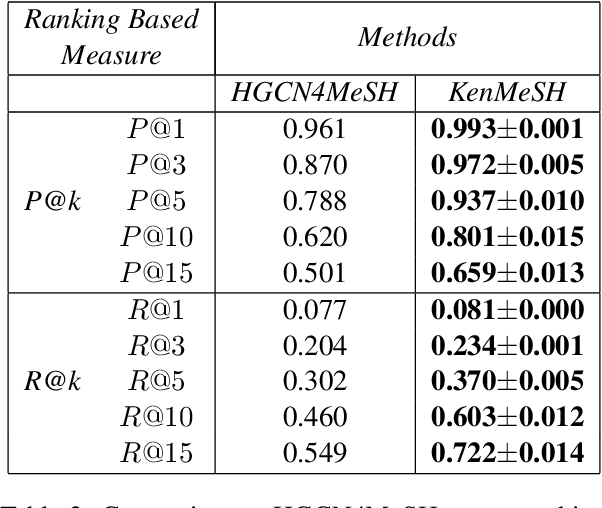
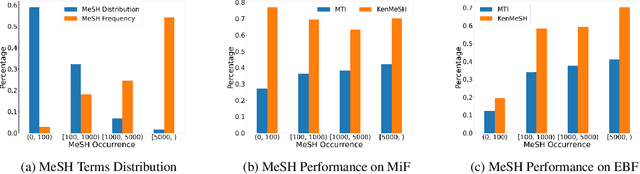
Currently, Medical Subject Headings (MeSH) are manually assigned to every biomedical article published and subsequently recorded in the PubMed database to facilitate retrieving relevant information. With the rapid growth of the PubMed database, large-scale biomedical document indexing becomes increasingly important. MeSH indexing is a challenging task for machine learning, as it needs to assign multiple labels to each article from an extremely large hierachically organized collection. To address this challenge, we propose KenMeSH, an end-to-end model that combines new text features and a dynamic \textbf{K}nowledge-\textbf{en}hanced mask attention that integrates document features with MeSH label hierarchy and journal correlation features to index MeSH terms. Experimental results show the proposed method achieves state-of-the-art performance on a number of measures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge