Robert E. Mercer
Graph-tree Fusion Model with Bidirectional Information Propagation for Long Document Classification
Oct 03, 2024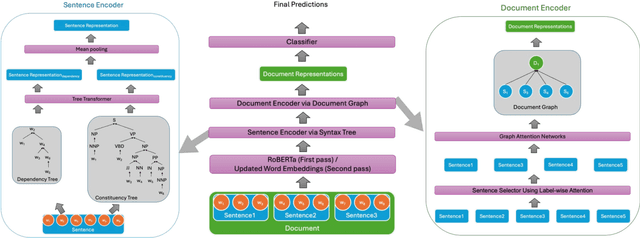
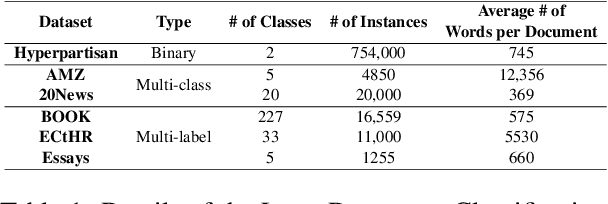
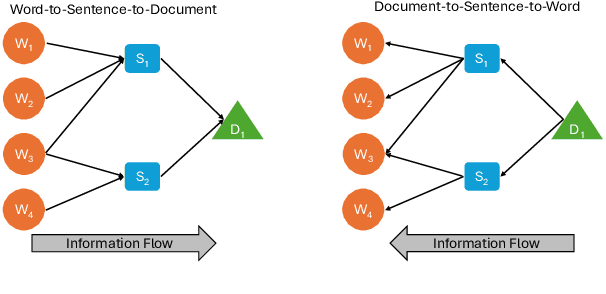
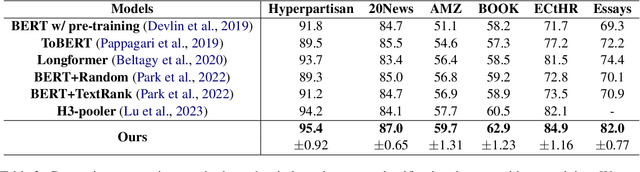
Abstract:Long document classification presents challenges in capturing both local and global dependencies due to their extensive content and complex structure. Existing methods often struggle with token limits and fail to adequately model hierarchical relationships within documents. To address these constraints, we propose a novel model leveraging a graph-tree structure. Our approach integrates syntax trees for sentence encodings and document graphs for document encodings, which capture fine-grained syntactic relationships and broader document contexts, respectively. We use Tree Transformers to generate sentence encodings, while a graph attention network models inter- and intra-sentence dependencies. During training, we implement bidirectional information propagation from word-to-sentence-to-document and vice versa, which enriches the contextual representation. Our proposed method enables a comprehensive understanding of content at all hierarchical levels and effectively handles arbitrarily long contexts without token limit constraints. Experimental results demonstrate the effectiveness of our approach in all types of long document classification tasks.
Auxiliary Knowledge-Induced Learning for Automatic Multi-Label Medical Document Classification
May 29, 2024Abstract:The International Classification of Diseases (ICD) is an authoritative medical classification system of different diseases and conditions for clinical and management purposes. ICD indexing assigns a subset of ICD codes to a medical record. Since human coding is labour-intensive and error-prone, many studies employ machine learning to automate the coding process. ICD coding is a challenging task, as it needs to assign multiple codes to each medical document from an extremely large hierarchically organized collection. In this paper, we propose a novel approach for ICD indexing that adopts three ideas: (1) we use a multi-level deep dilated residual convolution encoder to aggregate the information from the clinical notes and learn document representations across different lengths of the texts; (2) we formalize the task of ICD classification with auxiliary knowledge of the medical records, which incorporates not only the clinical texts but also different clinical code terminologies and drug prescriptions for better inferring the ICD codes; and (3) we introduce a graph convolutional network to leverage the co-occurrence patterns among ICD codes, aiming to enhance the quality of label representations. Experimental results show the proposed method achieves state-of-the-art performance on a number of measures.
Multi-stage Retrieve and Re-rank Model for Automatic Medical Coding Recommendation
May 29, 2024
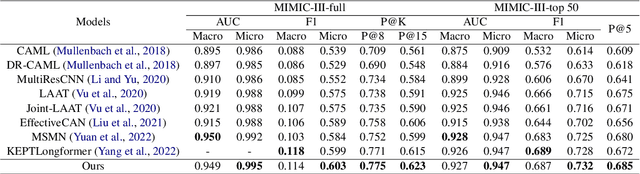
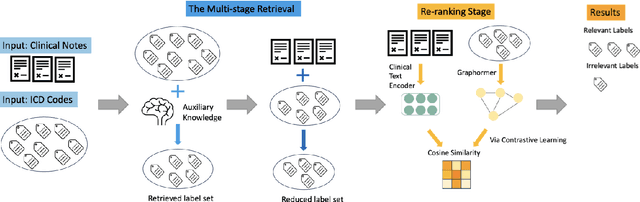

Abstract:The International Classification of Diseases (ICD) serves as a definitive medical classification system encompassing a wide range of diseases and conditions. The primary objective of ICD indexing is to allocate a subset of ICD codes to a medical record, which facilitates standardized documentation and management of various health conditions. Most existing approaches have suffered from selecting the proper label subsets from an extremely large ICD collection with a heavy long-tailed label distribution. In this paper, we leverage a multi-stage ``retrieve and re-rank'' framework as a novel solution to ICD indexing, via a hybrid discrete retrieval method, and re-rank retrieved candidates with contrastive learning that allows the model to make more accurate predictions from a simplified label space. The retrieval model is a hybrid of auxiliary knowledge of the electronic health records (EHR) and a discrete retrieval method (BM25), which efficiently collects high-quality candidates. In the last stage, we propose a label co-occurrence guided contrastive re-ranking model, which re-ranks the candidate labels by pulling together the clinical notes with positive ICD codes. Experimental results show the proposed method achieves state-of-the-art performance on a number of measures on the MIMIC-III benchmark.
Investigating the Learning Behaviour of In-context Learning: A Comparison with Supervised Learning
Aug 01, 2023Abstract:Large language models (LLMs) have shown remarkable capacity for in-context learning (ICL), where learning a new task from just a few training examples is done without being explicitly pre-trained. However, despite the success of LLMs, there has been little understanding of how ICL learns the knowledge from the given prompts. In this paper, to make progress toward understanding the learning behaviour of ICL, we train the same LLMs with the same demonstration examples via ICL and supervised learning (SL), respectively, and investigate their performance under label perturbations (i.e., noisy labels and label imbalance) on a range of classification tasks. First, via extensive experiments, we find that gold labels have significant impacts on the downstream in-context performance, especially for large language models; however, imbalanced labels matter little to ICL across all model sizes. Second, when comparing with SL, we show empirically that ICL is less sensitive to label perturbations than SL, and ICL gradually attains comparable performance to SL as the model size increases.
MeSHup: A Corpus for Full Text Biomedical Document Indexing
Apr 28, 2022
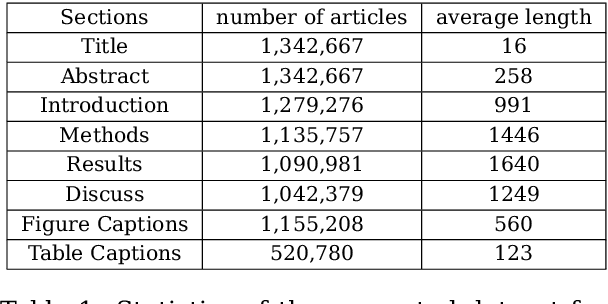
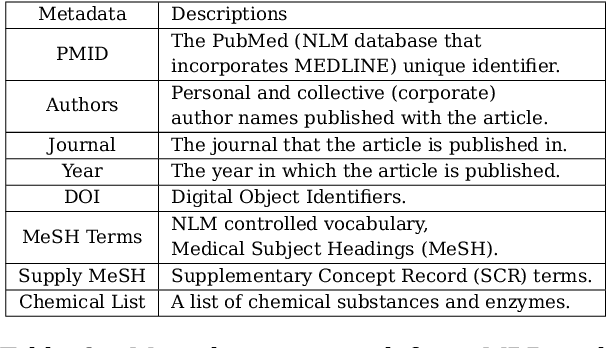
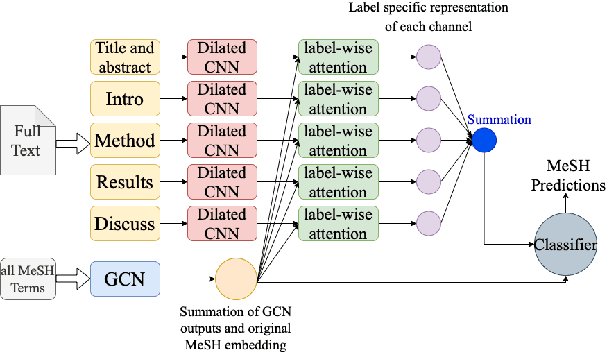
Abstract:Medical Subject Heading (MeSH) indexing refers to the problem of assigning a given biomedical document with the most relevant labels from an extremely large set of MeSH terms. Currently, the vast number of biomedical articles in the PubMed database are manually annotated by human curators, which is time consuming and costly; therefore, a computational system that can assist the indexing is highly valuable. When developing supervised MeSH indexing systems, the availability of a large-scale annotated text corpus is desirable. A publicly available, large corpus that permits robust evaluation and comparison of various systems is important to the research community. We release a large scale annotated MeSH indexing corpus, MeSHup, which contains 1,342,667 full text articles in English, together with the associated MeSH labels and metadata, authors, and publication venues that are collected from the MEDLINE database. We train an end-to-end model that combines features from documents and their associated labels on our corpus and report the new baseline.
Evaluation Benchmarks for Spanish Sentence Representations
Apr 15, 2022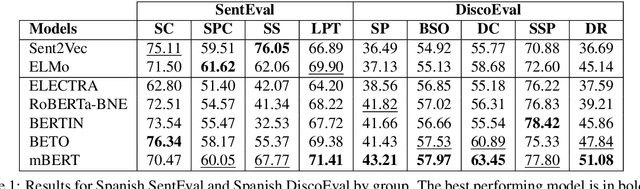
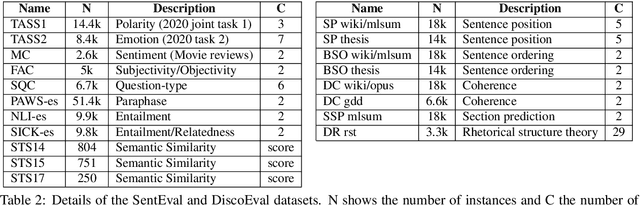
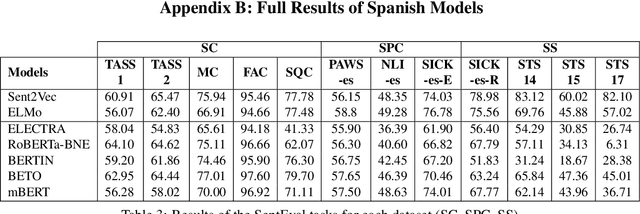
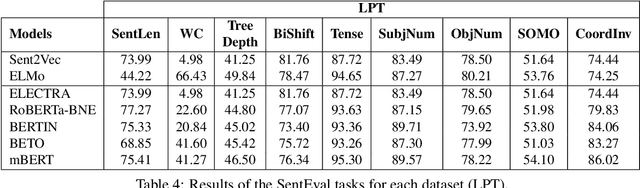
Abstract:Due to the success of pre-trained language models, versions of languages other than English have been released in recent years. This fact implies the need for resources to evaluate these models. In the case of Spanish, there are few ways to systematically assess the models' quality. In this paper, we narrow the gap by building two evaluation benchmarks. Inspired by previous work (Conneau and Kiela, 2018; Chen et al., 2019), we introduce Spanish SentEval and Spanish DiscoEval, aiming to assess the capabilities of stand-alone and discourse-aware sentence representations, respectively. Our benchmarks include considerable pre-existing and newly constructed datasets that address different tasks from various domains. In addition, we evaluate and analyze the most recent pre-trained Spanish language models to exhibit their capabilities and limitations. As an example, we discover that for the case of discourse evaluation tasks, mBERT, a language model trained on multiple languages, usually provides a richer latent representation than models trained only with documents in Spanish. We hope our contribution will motivate a fairer, more comparable, and less cumbersome way to evaluate future Spanish language models.
KenMeSH: Knowledge-enhanced End-to-end Biomedical Text Labelling
Mar 14, 2022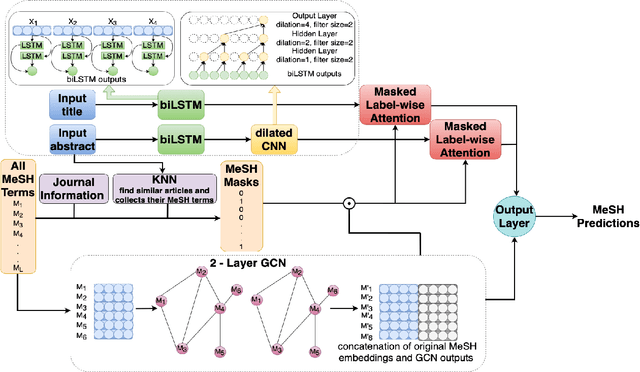
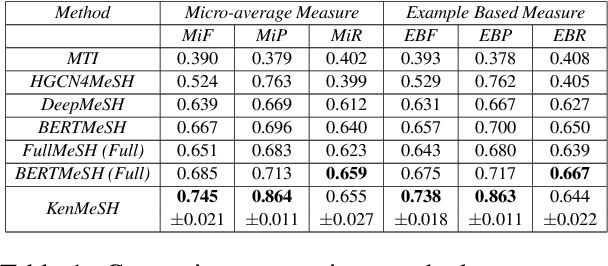
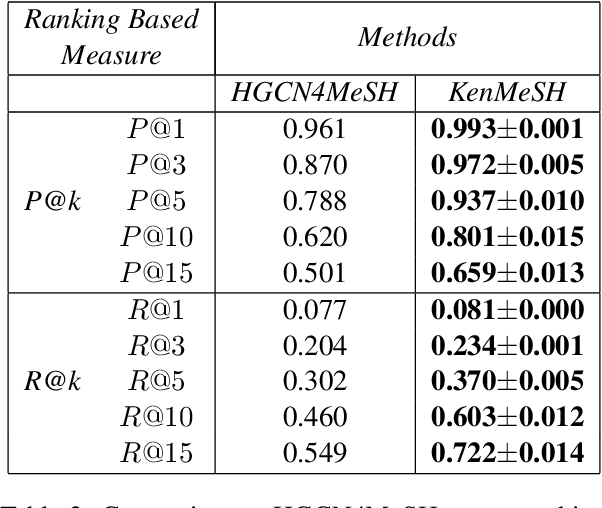
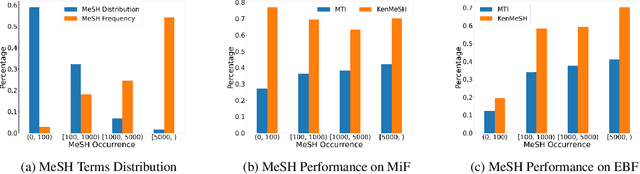
Abstract:Currently, Medical Subject Headings (MeSH) are manually assigned to every biomedical article published and subsequently recorded in the PubMed database to facilitate retrieving relevant information. With the rapid growth of the PubMed database, large-scale biomedical document indexing becomes increasingly important. MeSH indexing is a challenging task for machine learning, as it needs to assign multiple labels to each article from an extremely large hierachically organized collection. To address this challenge, we propose KenMeSH, an end-to-end model that combines new text features and a dynamic \textbf{K}nowledge-\textbf{en}hanced mask attention that integrates document features with MeSH label hierarchy and journal correlation features to index MeSH terms. Experimental results show the proposed method achieves state-of-the-art performance on a number of measures.
Improving Tree-LSTM with Tree Attention
Jan 01, 2019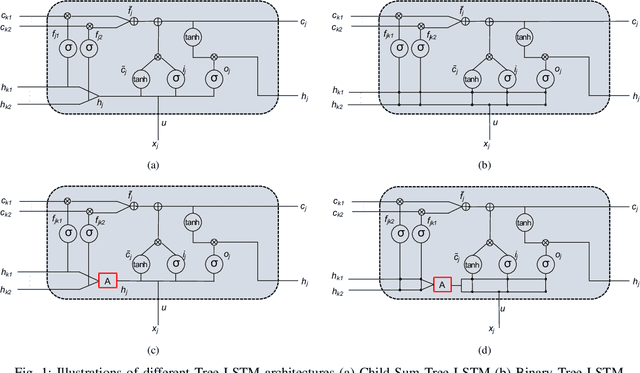
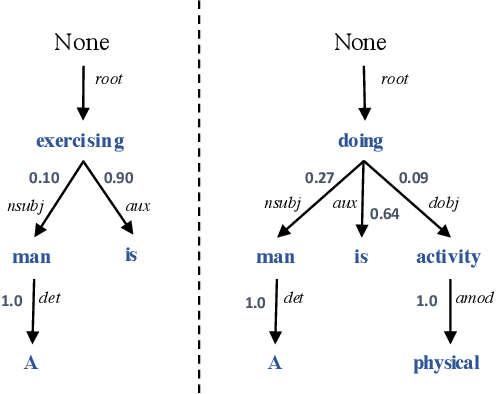
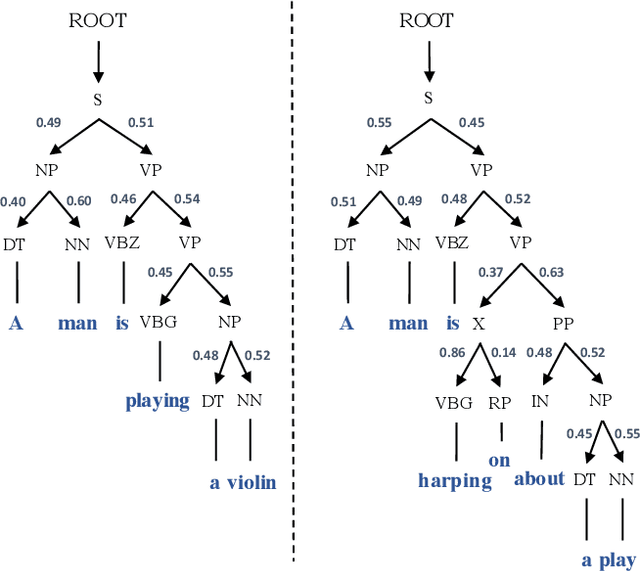
Abstract:In Natural Language Processing (NLP), we often need to extract information from tree topology. Sentence structure can be represented via a dependency tree or a constituency tree structure. For this reason, a variant of LSTMs, named Tree-LSTM, was proposed to work on tree topology. In this paper, we design a generalized attention framework for both dependency and constituency trees by encoding variants of decomposable attention inside a Tree-LSTM cell. We evaluated our models on a semantic relatedness task and achieved notable results compared to Tree-LSTM based methods with no attention as well as other neural and non-neural methods and good results compared to Tree-LSTM based methods with attention.
A Novel Neural Sequence Model with Multiple Attentions for Word Sense Disambiguation
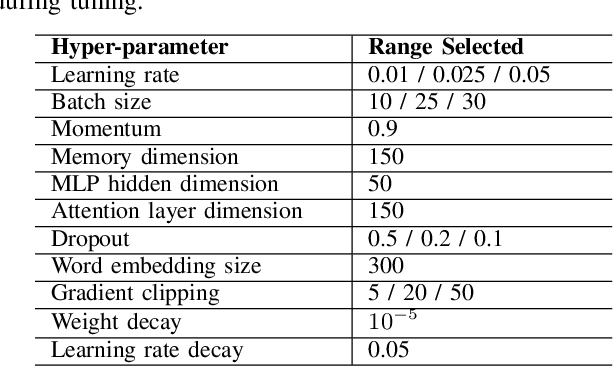
Sep 04, 2018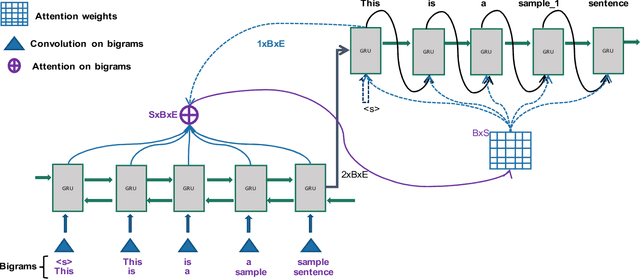
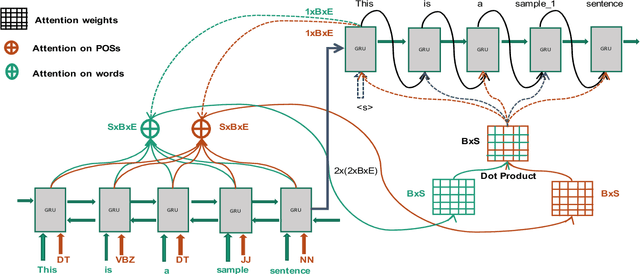
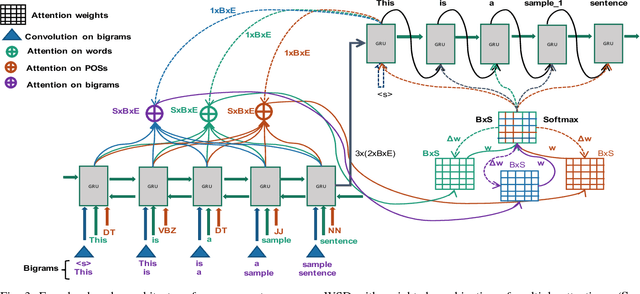
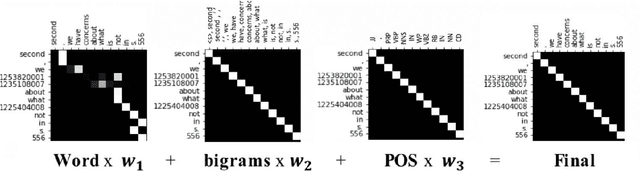
Abstract:Word sense disambiguation (WSD) is a well researched problem in computational linguistics. Different research works have approached this problem in different ways. Some state of the art results that have been achieved for this problem are by supervised models in terms of accuracy, but they often fall behind flexible knowledge-based solutions which use engineered features as well as human annotators to disambiguate every target word. This work focuses on bridging this gap using neural sequence models incorporating the well-known attention mechanism. The main gist of our work is to combine multiple attentions on different linguistic features through weights and to provide a unified framework for doing this. This weighted attention allows the model to easily disambiguate the sense of an ambiguous word by attending over a suitable portion of a sentence. Our extensive experiments show that multiple attention enables a more versatile encoder-decoder model leading to state of the art results.
Identifying Protein-Protein Interaction using Tree LSTM and Structured Attention
Jul 27, 2018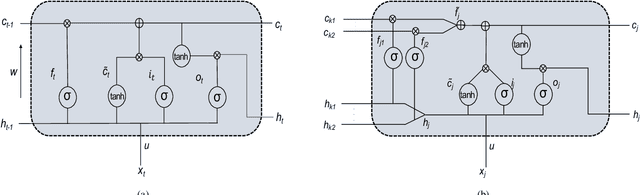
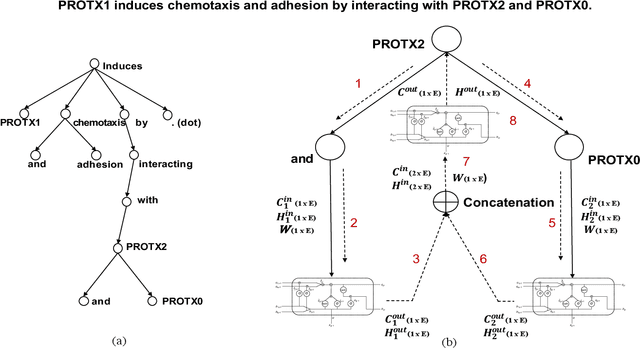
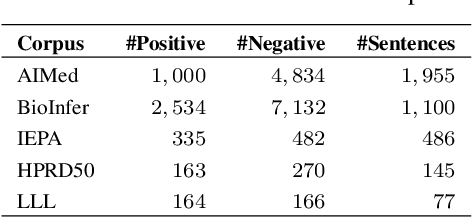
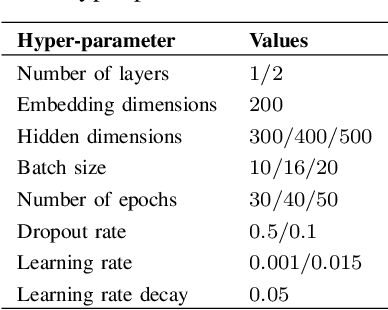
Abstract:Identifying interactions between proteins is important to understand underlying biological processes. Extracting a protein-protein interaction (PPI) from the raw text is often very difficult. Previous supervised learning methods have used handcrafted features on human-annotated data sets. In this paper, we propose a novel tree recurrent neural network with structured attention architecture for doing PPI. Our architecture achieves state of the art results (precision, recall, and F1-score) on the AIMed and BioInfer benchmark data sets. Moreover, our models achieve a significant improvement over previous best models without any explicit feature extraction. Our experimental results show that traditional recurrent networks have inferior performance compared to tree recurrent networks for the supervised PPI problem.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge