Yujuan Fu
DermaVQA-DAS: Dermatology Assessment Schema (DAS) & Datasets for Closed-Ended Question Answering & Segmentation in Patient-Generated Dermatology Images
Dec 30, 2025Abstract:Recent advances in dermatological image analysis have been driven by large-scale annotated datasets; however, most existing benchmarks focus on dermatoscopic images and lack patient-authored queries and clinical context, limiting their applicability to patient-centered care. To address this gap, we introduce DermaVQA-DAS, an extension of the DermaVQA dataset that supports two complementary tasks: closed-ended question answering (QA) and dermatological lesion segmentation. Central to this work is the Dermatology Assessment Schema (DAS), a novel expert-developed framework that systematically captures clinically meaningful dermatological features in a structured and standardized form. DAS comprises 36 high-level and 27 fine-grained assessment questions, with multiple-choice options in English and Chinese. Leveraging DAS, we provide expert-annotated datasets for both closed QA and segmentation and benchmark state-of-the-art multimodal models. For segmentation, we evaluate multiple prompting strategies and show that prompt design impacts performance: the default prompt achieves the best results under Mean-of-Max and Mean-of-Mean evaluation aggregation schemes, while an augmented prompt incorporating both patient query title and content yields the highest performance under majority-vote-based microscore evaluation, achieving a Jaccard index of 0.395 and a Dice score of 0.566 with BiomedParse. For closed-ended QA, overall performance is strong across models, with average accuracies ranging from 0.729 to 0.798; o3 achieves the best overall accuracy (0.798), closely followed by GPT-4.1 (0.796), while Gemini-1.5-Pro shows competitive performance within the Gemini family (0.783). We publicly release DermaVQA-DAS, the DAS schema, and evaluation protocols to support and accelerate future research in patient-centered dermatological vision-language modeling (https://osf.io/72rp3).
MEDEC: A Benchmark for Medical Error Detection and Correction in Clinical Notes
Dec 26, 2024Abstract:Several studies showed that Large Language Models (LLMs) can answer medical questions correctly, even outperforming the average human score in some medical exams. However, to our knowledge, no study has been conducted to assess the ability of language models to validate existing or generated medical text for correctness and consistency. In this paper, we introduce MEDEC (https://github.com/abachaa/MEDEC), the first publicly available benchmark for medical error detection and correction in clinical notes, covering five types of errors (Diagnosis, Management, Treatment, Pharmacotherapy, and Causal Organism). MEDEC consists of 3,848 clinical texts, including 488 clinical notes from three US hospital systems that were not previously seen by any LLM. The dataset has been used for the MEDIQA-CORR shared task to evaluate seventeen participating systems [Ben Abacha et al., 2024]. In this paper, we describe the data creation methods and we evaluate recent LLMs (e.g., o1-preview, GPT-4, Claude 3.5 Sonnet, and Gemini 2.0 Flash) for the tasks of detecting and correcting medical errors requiring both medical knowledge and reasoning capabilities. We also conducted a comparative study where two medical doctors performed the same task on the MEDEC test set. The results showed that MEDEC is a sufficiently challenging benchmark to assess the ability of models to validate existing or generated notes and to correct medical errors. We also found that although recent LLMs have a good performance in error detection and correction, they are still outperformed by medical doctors in these tasks. We discuss the potential factors behind this gap, the insights from our experiments, the limitations of current evaluation metrics, and share potential pointers for future research.
Does Data Contamination Detection Work (Well) for LLMs? A Survey and Evaluation on Detection Assumptions
Oct 24, 2024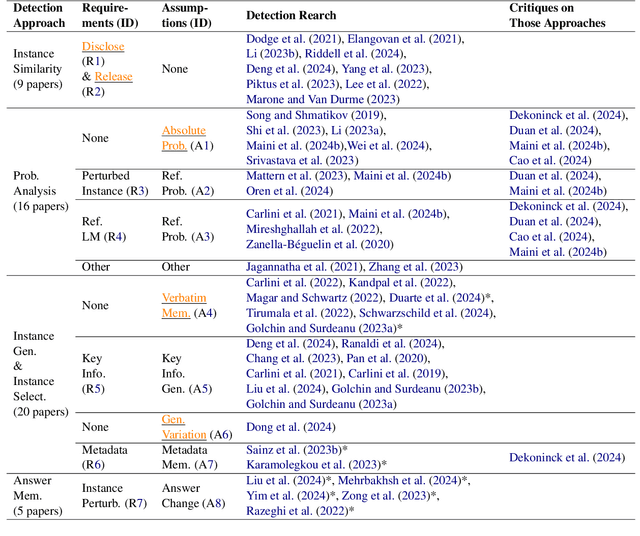
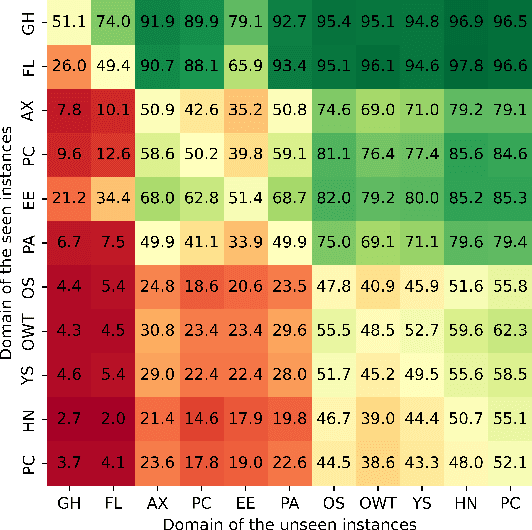
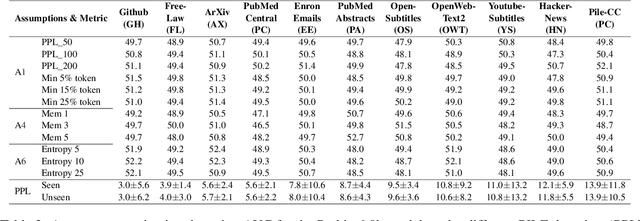
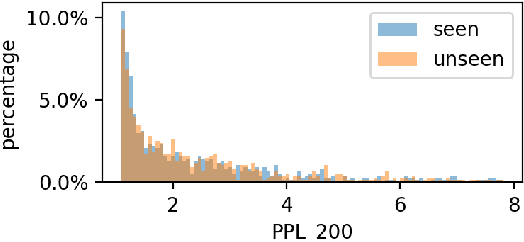
Abstract:Large language models (LLMs) have demonstrated great performance across various benchmarks, showing potential as general-purpose task solvers. However, as LLMs are typically trained on vast amounts of data, a significant concern in their evaluation is data contamination, where overlap between training data and evaluation datasets inflates performance assessments. While multiple approaches have been developed to identify data contamination, these approaches rely on specific assumptions that may not hold universally across different settings. To bridge this gap, we systematically review 47 papers on data contamination detection, categorize the underlying assumptions, and assess whether they have been rigorously validated. We identify and analyze eight categories of assumptions and test three of them as case studies. Our analysis reveals that when classifying instances used for pretraining LLMs, detection approaches based on these three assumptions perform close to random guessing, suggesting that current LLMs learn data distributions rather than memorizing individual instances. Overall, this work underscores the importance of approaches clearly stating their underlying assumptions and testing their validity across various scenarios.
CACER: Clinical Concept Annotations for Cancer Events and Relations
Sep 05, 2024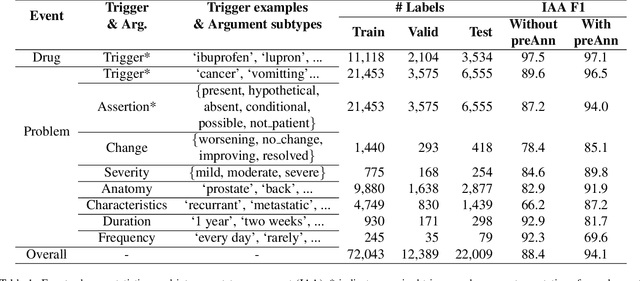
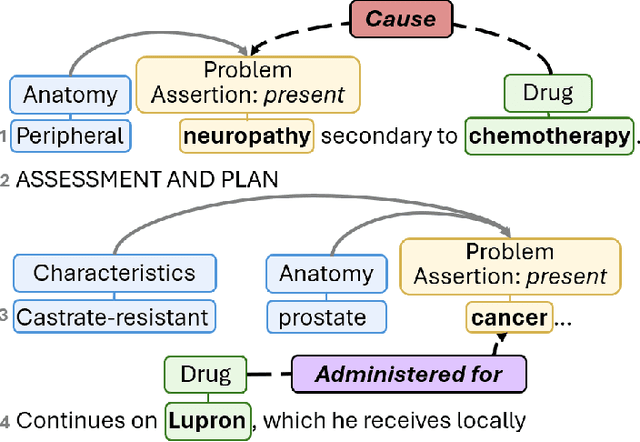
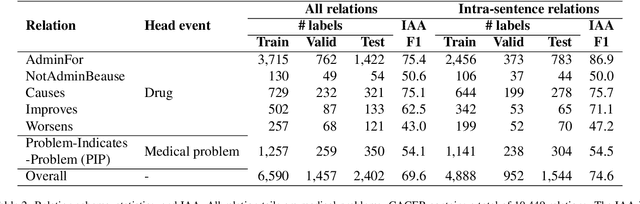
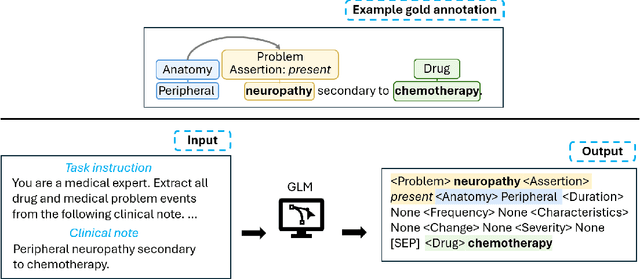
Abstract:Clinical notes contain unstructured representations of patient histories, including the relationships between medical problems and prescription drugs. To investigate the relationship between cancer drugs and their associated symptom burden, we extract structured, semantic representations of medical problem and drug information from the clinical narratives of oncology notes. We present Clinical Concept Annotations for Cancer Events and Relations (CACER), a novel corpus with fine-grained annotations for over 48,000 medical problems and drug events and 10,000 drug-problem and problem-problem relations. Leveraging CACER, we develop and evaluate transformer-based information extraction (IE) models such as BERT, Flan-T5, Llama3, and GPT-4 using fine-tuning and in-context learning (ICL). In event extraction, the fine-tuned BERT and Llama3 models achieved the highest performance at 88.2-88.0 F1, which is comparable to the inter-annotator agreement (IAA) of 88.4 F1. In relation extraction, the fine-tuned BERT, Flan-T5, and Llama3 achieved the highest performance at 61.8-65.3 F1. GPT-4 with ICL achieved the worst performance across both tasks. The fine-tuned models significantly outperformed GPT-4 in ICL, highlighting the importance of annotated training data and model optimization. Furthermore, the BERT models performed similarly to Llama3. For our task, LLMs offer no performance advantage over the smaller BERT models. The results emphasize the need for annotated training data to optimize models. Multiple fine-tuned transformer models achieved performance comparable to IAA for several extraction tasks.
* This is a pre-copy-editing, author-produced PDF of an article accepted for publication in JAMIA following peer review. The definitive publisher-authenticated version is available online at https://academic.oup.com/jamia/advance-article/doi/10.1093/jamia/ocae231/7748302
Extracting Social Determinants of Health from Pediatric Patient Notes Using Large Language Models: Novel Corpus and Methods
Apr 04, 2024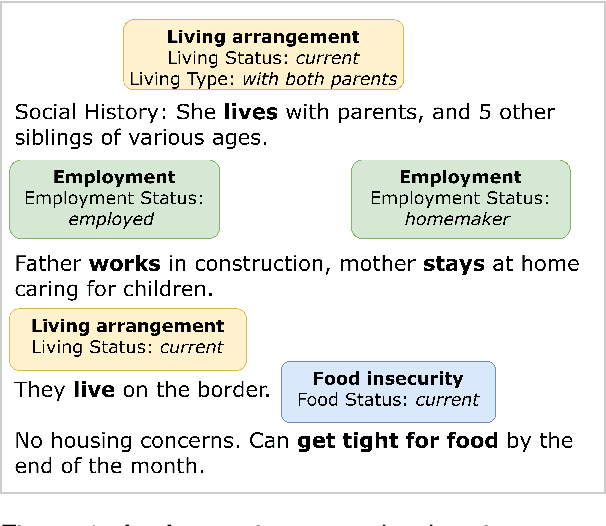
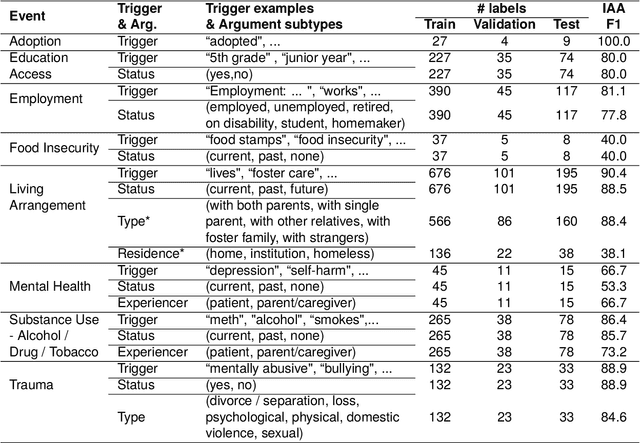
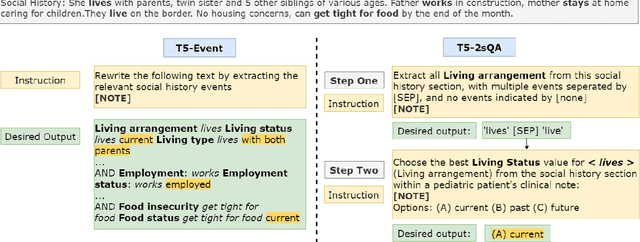
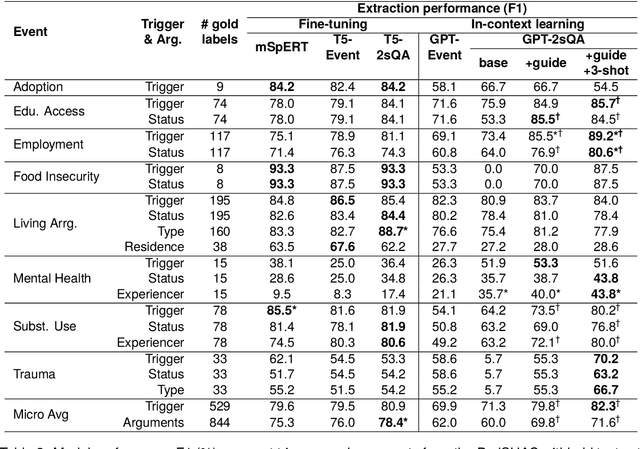
Abstract:Social determinants of health (SDoH) play a critical role in shaping health outcomes, particularly in pediatric populations where interventions can have long-term implications. SDoH are frequently studied in the Electronic Health Record (EHR), which provides a rich repository for diverse patient data. In this work, we present a novel annotated corpus, the Pediatric Social History Annotation Corpus (PedSHAC), and evaluate the automatic extraction of detailed SDoH representations using fine-tuned and in-context learning methods with Large Language Models (LLMs). PedSHAC comprises annotated social history sections from 1,260 clinical notes obtained from pediatric patients within the University of Washington (UW) hospital system. Employing an event-based annotation scheme, PedSHAC captures ten distinct health determinants to encompass living and economic stability, prior trauma, education access, substance use history, and mental health with an overall annotator agreement of 81.9 F1. Our proposed fine-tuning LLM-based extractors achieve high performance at 78.4 F1 for event arguments. In-context learning approaches with GPT-4 demonstrate promise for reliable SDoH extraction with limited annotated examples, with extraction performance at 82.3 F1 for event triggers.
Prompt-based Extraction of Social Determinants of Health Using Few-shot Learning
Jun 12, 2023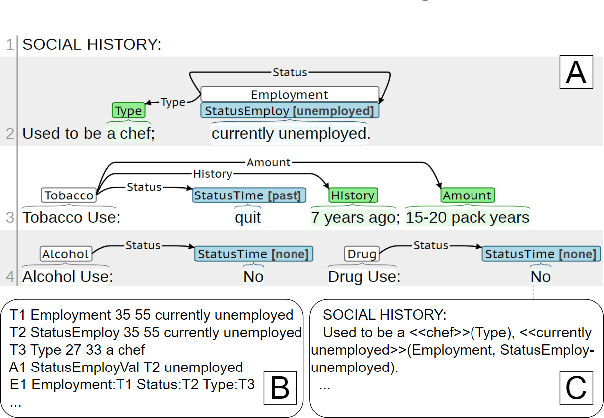
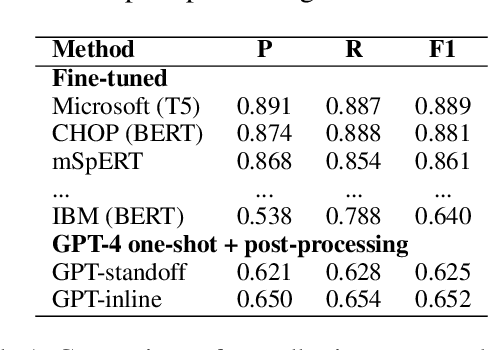
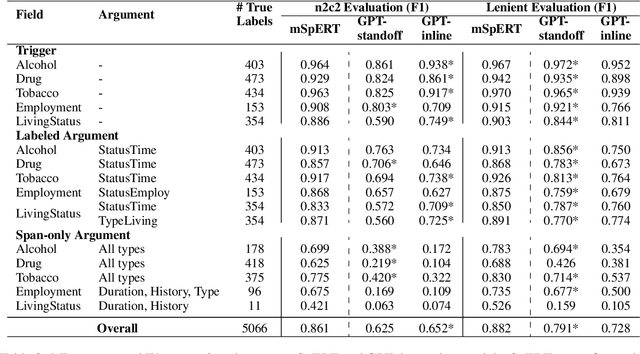
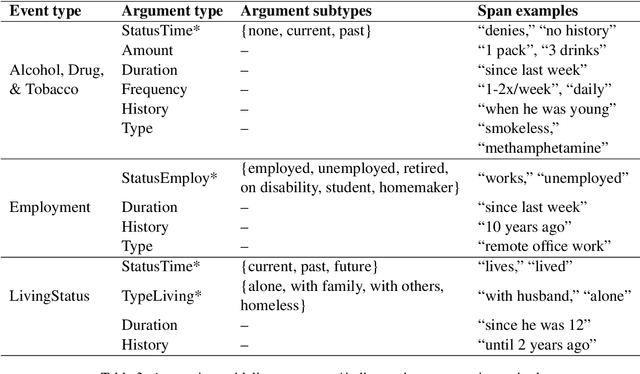
Abstract:Social determinants of health (SDOH) documented in the electronic health record through unstructured text are increasingly being studied to understand how SDOH impacts patient health outcomes. In this work, we utilize the Social History Annotation Corpus (SHAC), a multi-institutional corpus of de-identified social history sections annotated for SDOH, including substance use, employment, and living status information. We explore the automatic extraction of SDOH information with SHAC in both standoff and inline annotation formats using GPT-4 in a one-shot prompting setting. We compare GPT-4 extraction performance with a high-performing supervised approach and perform thorough error analyses. Our prompt-based GPT-4 method achieved an overall 0.652 F1 on the SHAC test set, similar to the 7th best-performing system among all teams in the n2c2 challenge with SHAC.
ACI-BENCH: a Novel Ambient Clinical Intelligence Dataset for Benchmarking Automatic Visit Note Generation
Jun 03, 2023Abstract:Recent immense breakthroughs in generative models such as in GPT4 have precipitated re-imagined ubiquitous usage of these models in all applications. One area that can benefit by improvements in artificial intelligence (AI) is healthcare. The note generation task from doctor-patient encounters, and its associated electronic medical record documentation, is one of the most arduous time-consuming tasks for physicians. It is also a natural prime potential beneficiary to advances in generative models. However with such advances, benchmarking is more critical than ever. Whether studying model weaknesses or developing new evaluation metrics, shared open datasets are an imperative part of understanding the current state-of-the-art. Unfortunately as clinic encounter conversations are not routinely recorded and are difficult to ethically share due to patient confidentiality, there are no sufficiently large clinic dialogue-note datasets to benchmark this task. Here we present the Ambient Clinical Intelligence Benchmark (ACI-BENCH) corpus, the largest dataset to date tackling the problem of AI-assisted note generation from visit dialogue. We also present the benchmark performances of several common state-of-the-art approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge