Kevin Lybarger
Advancing AI Trustworthiness Through Patient Simulation: Risk Assessment of Conversational Agents for Antidepressant Selection
Feb 11, 2026Abstract:Objective: This paper introduces a patient simulator designed to enable scalable, automated evaluation of healthcare conversational agents. The simulator generates realistic, controllable patient interactions that systematically vary across medical, linguistic, and behavioral dimensions, allowing annotators and an independent AI judge to assess agent performance, identify hallucinations and inaccuracies, and characterize risk patterns across diverse patient populations. Methods: The simulator is grounded in the NIST AI Risk Management Framework and integrates three profile components reflecting different dimensions of patient variation: (1) medical profiles constructed from electronic health records in the All of Us Research Program; (2) linguistic profiles modeling variation in health literacy and condition-specific communication patterns; and (3) behavioral profiles representing empirically observed interaction patterns, including cooperation, distraction, and adversarial engagement. We evaluated the simulator's effectiveness in identifying errors in an AI decision aid for antidepressant selection. Results: We generated 500 conversations between the patient simulator and the AI decision aid across systematic combinations of five linguistic and three behavioral profiles. Human annotators assessed 1,787 medical concepts across 100 conversations, achieving high agreement (F1=0.94, \k{appa}=0.73), and the LLM judge achieved comparable agreement with human annotators (F1=0.94, \k{appa}=0.78; paired bootstrap p=0.21). The simulator revealed a monotonic degradation in AI decision aid performance across the health literacy spectrum: rank-one concept retrieval accuracy increased from 47.9% for limited health literacy to 69.1% for functional and 81.6% for proficient.
DF-RAG: Query-Aware Diversity for Retrieval-Augmented Generation
Jan 23, 2026Abstract:Retrieval-augmented generation (RAG) is a common technique for grounding language model outputs in domain-specific information. However, RAG is often challenged by reasoning-intensive question-answering (QA), since common retrieval methods like cosine similarity maximize relevance at the cost of introducing redundant content, which can reduce information recall. To address this, we introduce Diversity-Focused Retrieval-Augmented Generation (DF-RAG), which systematically incorporates diversity into the retrieval step to improve performance on complex, reasoning-intensive QA benchmarks. DF-RAG builds upon the Maximal Marginal Relevance framework to select information chunks that are both relevant to the query and maximally dissimilar from each other. A key innovation of DF-RAG is its ability to optimize the level of diversity for each query dynamically at test time without requiring any additional fine-tuning or prior information. We show that DF-RAG improves F1 performance on reasoning-intensive QA benchmarks by 4-10 percent over vanilla RAG using cosine similarity and also outperforms other established baselines. Furthermore, we estimate an Oracle ceiling of up to 18 percent absolute F1 gains over vanilla RAG, of which DF-RAG captures up to 91.3 percent.
Identifying Imaging Follow-Up in Radiology Reports: A Comparative Analysis of Traditional ML and LLM Approaches
Nov 14, 2025Abstract:Large language models (LLMs) have shown considerable promise in clinical natural language processing, yet few domain-specific datasets exist to rigorously evaluate their performance on radiology tasks. In this work, we introduce an annotated corpus of 6,393 radiology reports from 586 patients, each labeled for follow-up imaging status, to support the development and benchmarking of follow-up adherence detection systems. Using this corpus, we systematically compared traditional machine-learning classifiers, including logistic regression (LR), support vector machines (SVM), Longformer, and a fully fine-tuned Llama3-8B-Instruct, with recent generative LLMs. To evaluate generative LLMs, we tested GPT-4o and the open-source GPT-OSS-20B under two configurations: a baseline (Base) and a task-optimized (Advanced) setting that focused inputs on metadata, recommendation sentences, and their surrounding context. A refined prompt for GPT-OSS-20B further improved reasoning accuracy. Performance was assessed using precision, recall, and F1 scores with 95% confidence intervals estimated via non-parametric bootstrapping. Inter-annotator agreement was high (F1 = 0.846). GPT-4o (Advanced) achieved the best performance (F1 = 0.832), followed closely by GPT-OSS-20B (Advanced; F1 = 0.828). LR and SVM also performed strongly (F1 = 0.776 and 0.775), underscoring that while LLMs approach human-level agreement through prompt optimization, interpretable and resource-efficient models remain valuable baselines.
Towards AI-Driven Human-Machine Co-Teaming for Adaptive and Agile Cyber Security Operation Centers
May 09, 2025Abstract:Security Operations Centers (SOCs) face growing challenges in managing cybersecurity threats due to an overwhelming volume of alerts, a shortage of skilled analysts, and poorly integrated tools. Human-AI collaboration offers a promising path to augment the capabilities of SOC analysts while reducing their cognitive overload. To this end, we introduce an AI-driven human-machine co-teaming paradigm that leverages large language models (LLMs) to enhance threat intelligence, alert triage, and incident response workflows. We present a vision in which LLM-based AI agents learn from human analysts the tacit knowledge embedded in SOC operations, enabling the AI agents to improve their performance on SOC tasks through this co-teaming. We invite SOCs to collaborate with us to further develop this process and uncover replicable patterns where human-AI co-teaming yields measurable improvements in SOC productivity.
Adapting Biomedical Abstracts into Plain language using Large Language Models
Jan 26, 2025



Abstract:A vast amount of medical knowledge is available for public use through online health forums, and question-answering platforms on social media. The majority of the population in the United States doesn't have the right amount of health literacy to make the best use of that information. Health literacy means the ability to obtain and comprehend the basic health information to make appropriate health decisions. To build the bridge between this gap, organizations advocate adapting this medical knowledge into plain language. Building robust systems to automate the adaptations helps both medical and non-medical professionals best leverage the available information online. The goal of the Plain Language Adaptation of Biomedical Abstracts (PLABA) track is to adapt the biomedical abstracts in English language extracted from PubMed based on the questions asked in MedlinePlus for the general public using plain language at the sentence level. As part of this track, we leveraged the best open-source Large Language Models suitable and fine-tuned for dialog use cases. We compare and present the results for all of our systems and our ranking among the other participants' submissions. Our top performing GPT-4 based model ranked first in the avg. simplicity measure and 3rd on the avg. accuracy measure.
BioMistral-NLU: Towards More Generalizable Medical Language Understanding through Instruction Tuning
Oct 24, 2024



Abstract:Large language models (LLMs) such as ChatGPT are fine-tuned on large and diverse instruction-following corpora, and can generalize to new tasks. However, those instruction-tuned LLMs often perform poorly in specialized medical natural language understanding (NLU) tasks that require domain knowledge, granular text comprehension, and structured data extraction. To bridge the gap, we: (1) propose a unified prompting format for 7 important NLU tasks, % through span extraction and multi-choice question-answering (QA), (2) curate an instruction-tuning dataset, MNLU-Instruct, utilizing diverse existing open-source medical NLU corpora, and (3) develop BioMistral-NLU, a generalizable medical NLU model, through fine-tuning BioMistral on MNLU-Instruct. We evaluate BioMistral-NLU in a zero-shot setting, across 6 important NLU tasks, from two widely adopted medical NLU benchmarks: Biomedical Language Understanding Evaluation (BLUE) and Biomedical Language Understanding and Reasoning Benchmark (BLURB). Our experiments show that our BioMistral-NLU outperforms the original BioMistral, as well as the proprietary LLMs - ChatGPT and GPT-4. Our dataset-agnostic prompting strategy and instruction tuning step over diverse NLU tasks enhance LLMs' generalizability across diverse medical NLU tasks. Our ablation experiments show that instruction-tuning on a wider variety of tasks, even when the total number of training instances remains constant, enhances downstream zero-shot generalization.
CACER: Clinical Concept Annotations for Cancer Events and Relations
Sep 05, 2024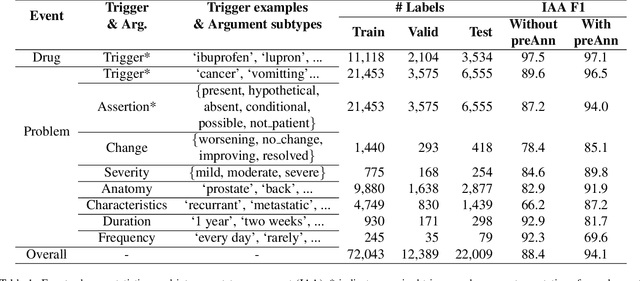
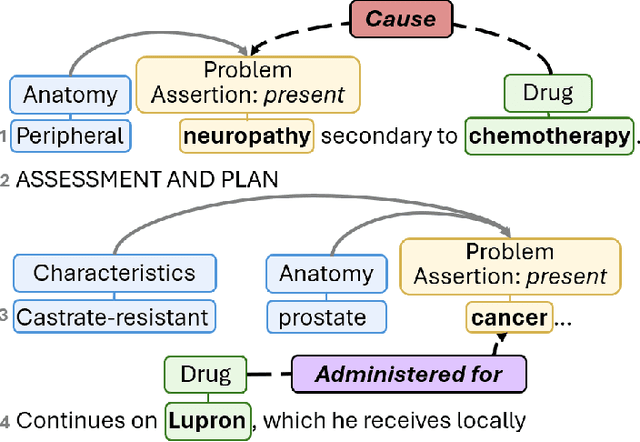
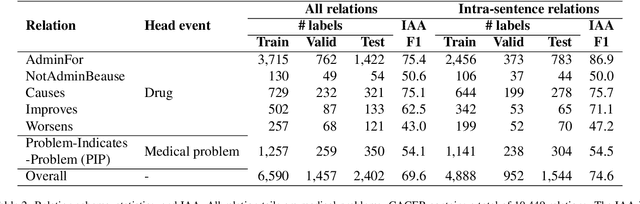
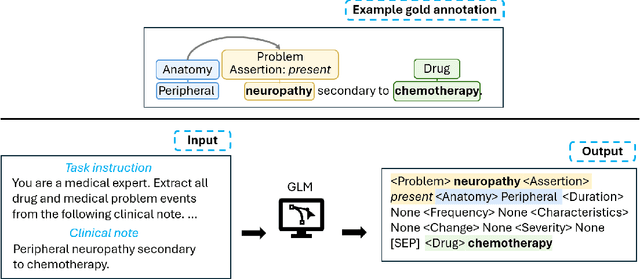
Abstract:Clinical notes contain unstructured representations of patient histories, including the relationships between medical problems and prescription drugs. To investigate the relationship between cancer drugs and their associated symptom burden, we extract structured, semantic representations of medical problem and drug information from the clinical narratives of oncology notes. We present Clinical Concept Annotations for Cancer Events and Relations (CACER), a novel corpus with fine-grained annotations for over 48,000 medical problems and drug events and 10,000 drug-problem and problem-problem relations. Leveraging CACER, we develop and evaluate transformer-based information extraction (IE) models such as BERT, Flan-T5, Llama3, and GPT-4 using fine-tuning and in-context learning (ICL). In event extraction, the fine-tuned BERT and Llama3 models achieved the highest performance at 88.2-88.0 F1, which is comparable to the inter-annotator agreement (IAA) of 88.4 F1. In relation extraction, the fine-tuned BERT, Flan-T5, and Llama3 achieved the highest performance at 61.8-65.3 F1. GPT-4 with ICL achieved the worst performance across both tasks. The fine-tuned models significantly outperformed GPT-4 in ICL, highlighting the importance of annotated training data and model optimization. Furthermore, the BERT models performed similarly to Llama3. For our task, LLMs offer no performance advantage over the smaller BERT models. The results emphasize the need for annotated training data to optimize models. Multiple fine-tuned transformer models achieved performance comparable to IAA for several extraction tasks.
* This is a pre-copy-editing, author-produced PDF of an article accepted for publication in JAMIA following peer review. The definitive publisher-authenticated version is available online at https://academic.oup.com/jamia/advance-article/doi/10.1093/jamia/ocae231/7748302
Classifying Human-Generated and AI-Generated Election Claims in Social Media
Apr 26, 2024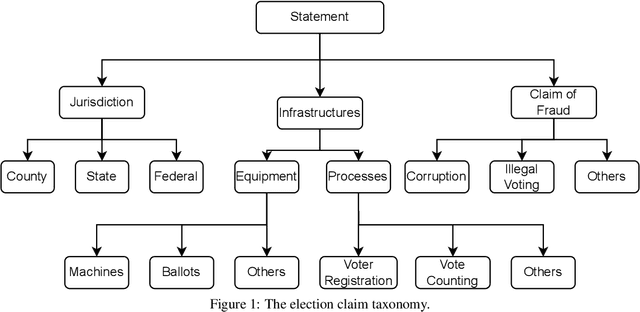
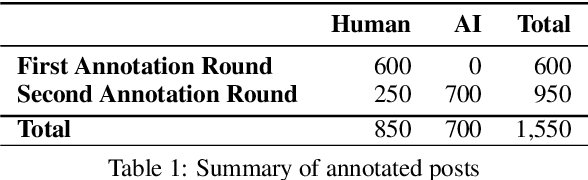

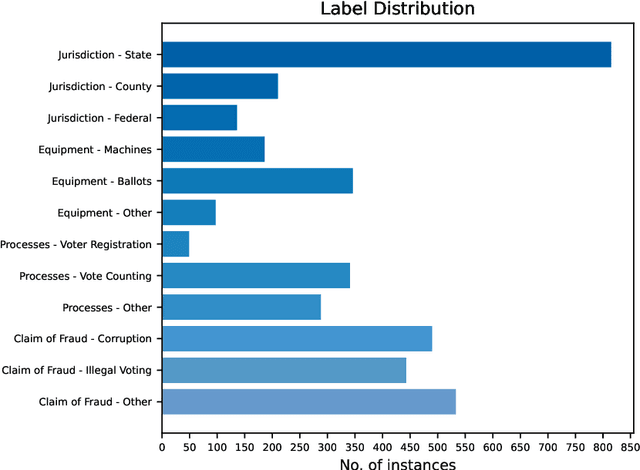
Abstract:Politics is one of the most prevalent topics discussed on social media platforms, particularly during major election cycles, where users engage in conversations about candidates and electoral processes. Malicious actors may use this opportunity to disseminate misinformation to undermine trust in the electoral process. The emergence of Large Language Models (LLMs) exacerbates this issue by enabling malicious actors to generate misinformation at an unprecedented scale. Artificial intelligence (AI)-generated content is often indistinguishable from authentic user content, raising concerns about the integrity of information on social networks. In this paper, we present a novel taxonomy for characterizing election-related claims. This taxonomy provides an instrument for analyzing election-related claims, with granular categories related to jurisdiction, equipment, processes, and the nature of claims. We introduce ElectAI, a novel benchmark dataset that consists of 9,900 tweets, each labeled as human- or AI-generated. For AI-generated tweets, the specific LLM variant that produced them is specified. We annotated a subset of 1,550 tweets using the proposed taxonomy to capture the characteristics of election-related claims. We explored the capabilities of LLMs in extracting the taxonomy attributes and trained various machine learning models using ElectAI to distinguish between human- and AI-generated posts and identify the specific LLM variant.
Extracting Social Determinants of Health from Pediatric Patient Notes Using Large Language Models: Novel Corpus and Methods
Apr 04, 2024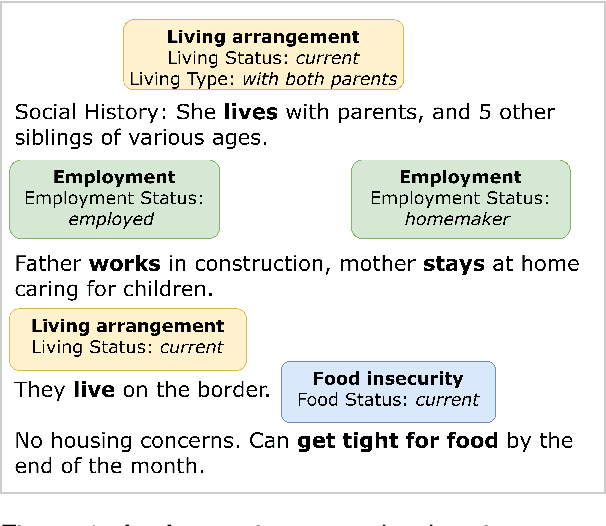
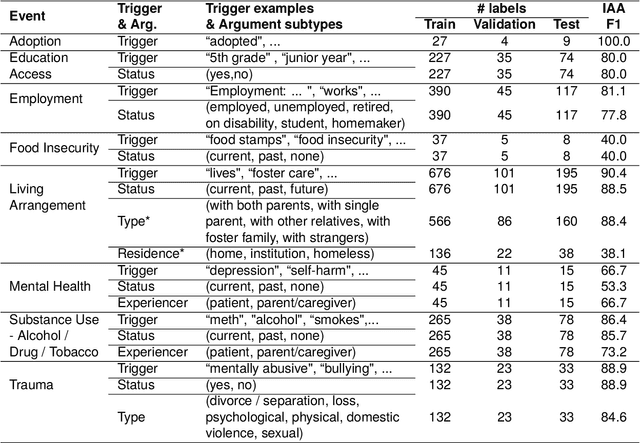
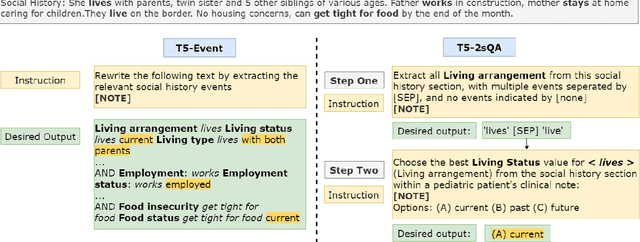
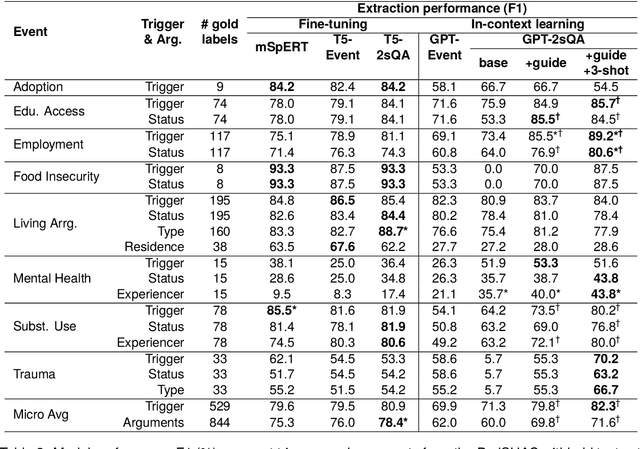
Abstract:Social determinants of health (SDoH) play a critical role in shaping health outcomes, particularly in pediatric populations where interventions can have long-term implications. SDoH are frequently studied in the Electronic Health Record (EHR), which provides a rich repository for diverse patient data. In this work, we present a novel annotated corpus, the Pediatric Social History Annotation Corpus (PedSHAC), and evaluate the automatic extraction of detailed SDoH representations using fine-tuned and in-context learning methods with Large Language Models (LLMs). PedSHAC comprises annotated social history sections from 1,260 clinical notes obtained from pediatric patients within the University of Washington (UW) hospital system. Employing an event-based annotation scheme, PedSHAC captures ten distinct health determinants to encompass living and economic stability, prior trauma, education access, substance use history, and mental health with an overall annotator agreement of 81.9 F1. Our proposed fine-tuning LLM-based extractors achieve high performance at 78.4 F1 for event arguments. In-context learning approaches with GPT-4 demonstrate promise for reliable SDoH extraction with limited annotated examples, with extraction performance at 82.3 F1 for event triggers.
A Novel Corpus of Annotated Medical Imaging Reports and Information Extraction Results Using BERT-based Language Models
Mar 27, 2024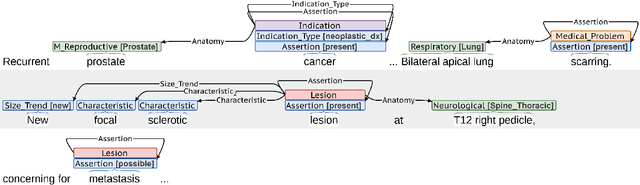
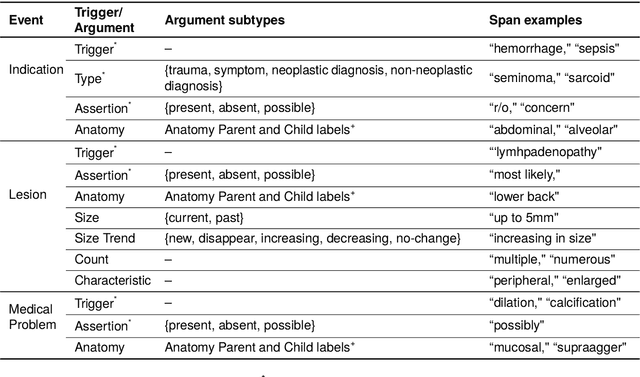
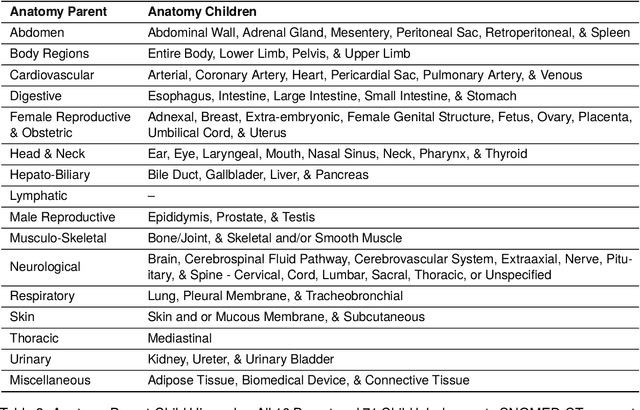
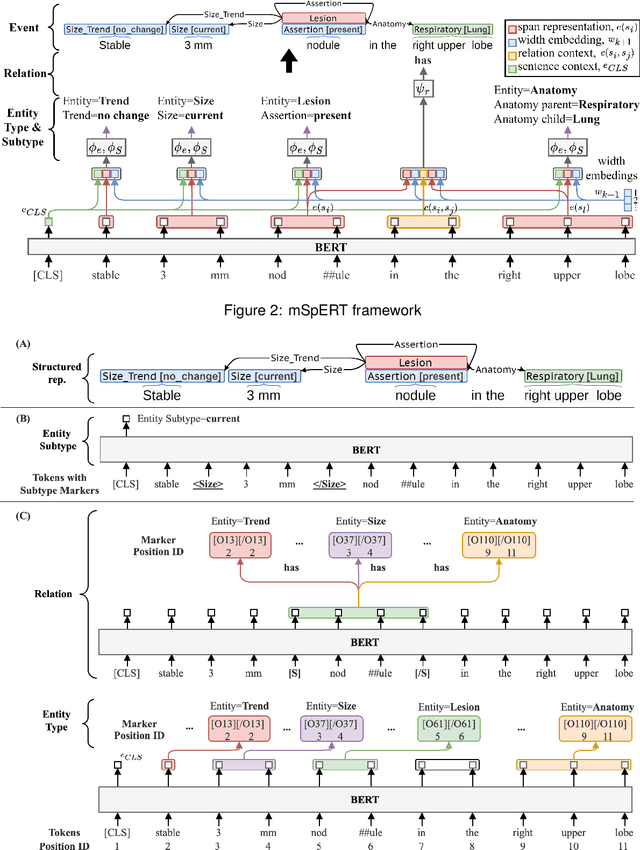
Abstract:Medical imaging is critical to the diagnosis, surveillance, and treatment of many health conditions, including oncological, neurological, cardiovascular, and musculoskeletal disorders, among others. Radiologists interpret these complex, unstructured images and articulate their assessments through narrative reports that remain largely unstructured. This unstructured narrative must be converted into a structured semantic representation to facilitate secondary applications such as retrospective analyses or clinical decision support. Here, we introduce the Corpus of Annotated Medical Imaging Reports (CAMIR), which includes 609 annotated radiology reports from three imaging modality types: Computed Tomography, Magnetic Resonance Imaging, and Positron Emission Tomography-Computed Tomography. Reports were annotated using an event-based schema that captures clinical indications, lesions, and medical problems. Each event consists of a trigger and multiple arguments, and a majority of the argument types, including anatomy, normalize the spans to pre-defined concepts to facilitate secondary use. CAMIR uniquely combines a granular event structure and concept normalization. To extract CAMIR events, we explored two BERT (Bi-directional Encoder Representation from Transformers)-based architectures, including an existing architecture (mSpERT) that jointly extracts all event information and a multi-step approach (PL-Marker++) that we augmented for the CAMIR schema.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge