Özlem Uzuner
Adapting Biomedical Abstracts into Plain language using Large Language Models
Jan 26, 2025



Abstract:A vast amount of medical knowledge is available for public use through online health forums, and question-answering platforms on social media. The majority of the population in the United States doesn't have the right amount of health literacy to make the best use of that information. Health literacy means the ability to obtain and comprehend the basic health information to make appropriate health decisions. To build the bridge between this gap, organizations advocate adapting this medical knowledge into plain language. Building robust systems to automate the adaptations helps both medical and non-medical professionals best leverage the available information online. The goal of the Plain Language Adaptation of Biomedical Abstracts (PLABA) track is to adapt the biomedical abstracts in English language extracted from PubMed based on the questions asked in MedlinePlus for the general public using plain language at the sentence level. As part of this track, we leveraged the best open-source Large Language Models suitable and fine-tuned for dialog use cases. We compare and present the results for all of our systems and our ranking among the other participants' submissions. Our top performing GPT-4 based model ranked first in the avg. simplicity measure and 3rd on the avg. accuracy measure.
CACER: Clinical Concept Annotations for Cancer Events and Relations
Sep 05, 2024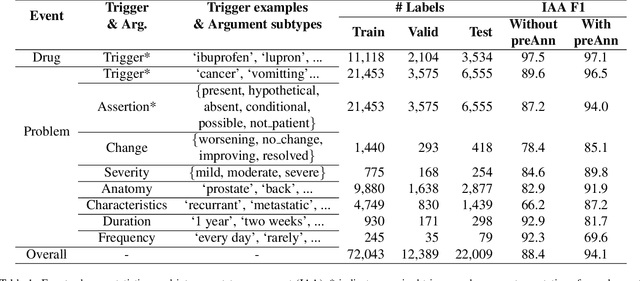
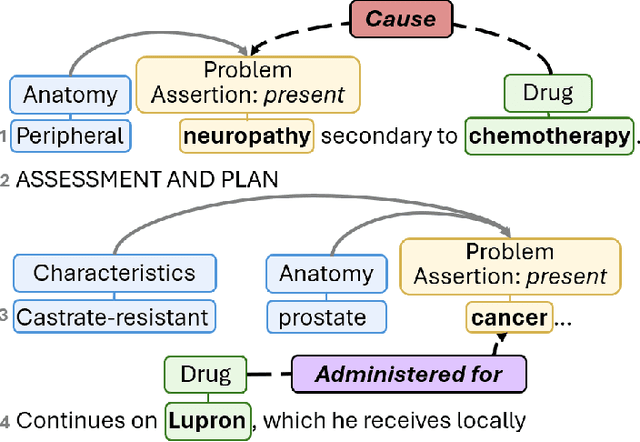
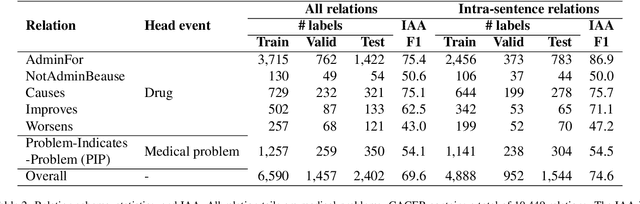
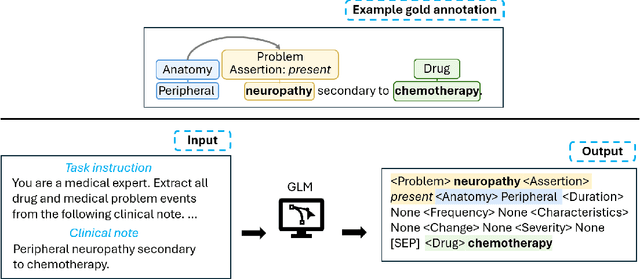
Abstract:Clinical notes contain unstructured representations of patient histories, including the relationships between medical problems and prescription drugs. To investigate the relationship between cancer drugs and their associated symptom burden, we extract structured, semantic representations of medical problem and drug information from the clinical narratives of oncology notes. We present Clinical Concept Annotations for Cancer Events and Relations (CACER), a novel corpus with fine-grained annotations for over 48,000 medical problems and drug events and 10,000 drug-problem and problem-problem relations. Leveraging CACER, we develop and evaluate transformer-based information extraction (IE) models such as BERT, Flan-T5, Llama3, and GPT-4 using fine-tuning and in-context learning (ICL). In event extraction, the fine-tuned BERT and Llama3 models achieved the highest performance at 88.2-88.0 F1, which is comparable to the inter-annotator agreement (IAA) of 88.4 F1. In relation extraction, the fine-tuned BERT, Flan-T5, and Llama3 achieved the highest performance at 61.8-65.3 F1. GPT-4 with ICL achieved the worst performance across both tasks. The fine-tuned models significantly outperformed GPT-4 in ICL, highlighting the importance of annotated training data and model optimization. Furthermore, the BERT models performed similarly to Llama3. For our task, LLMs offer no performance advantage over the smaller BERT models. The results emphasize the need for annotated training data to optimize models. Multiple fine-tuned transformer models achieved performance comparable to IAA for several extraction tasks.
* This is a pre-copy-editing, author-produced PDF of an article accepted for publication in JAMIA following peer review. The definitive publisher-authenticated version is available online at https://academic.oup.com/jamia/advance-article/doi/10.1093/jamia/ocae231/7748302
Prompt-based Extraction of Social Determinants of Health Using Few-shot Learning
Jun 12, 2023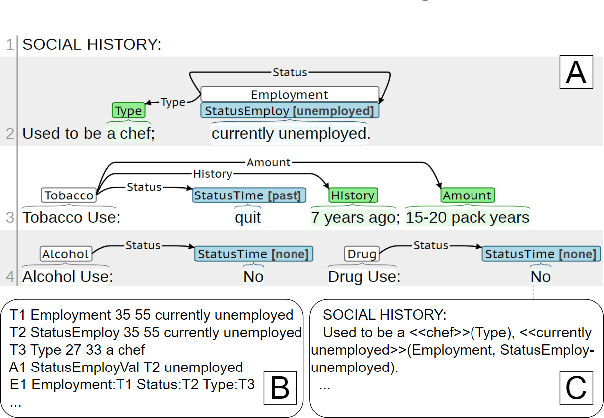
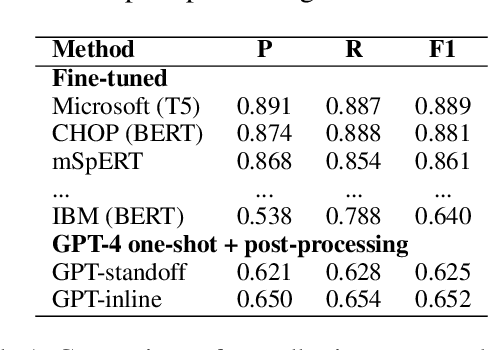
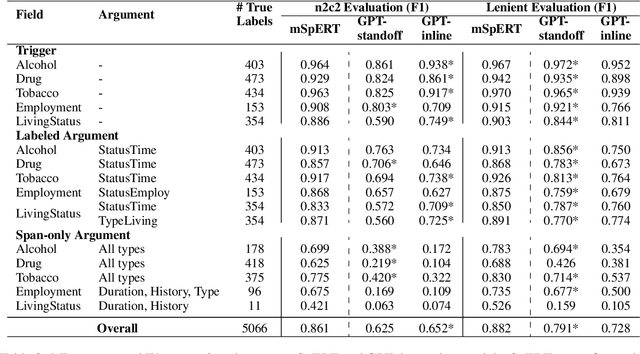
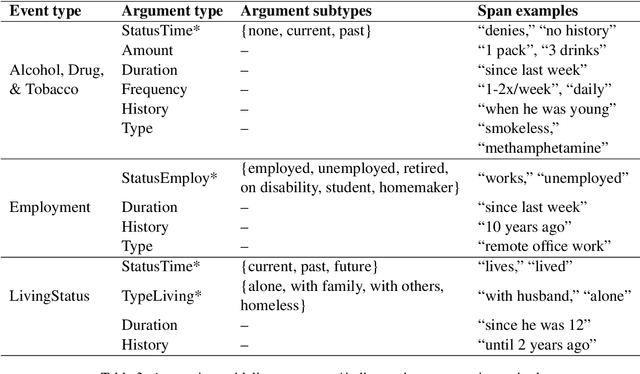
Abstract:Social determinants of health (SDOH) documented in the electronic health record through unstructured text are increasingly being studied to understand how SDOH impacts patient health outcomes. In this work, we utilize the Social History Annotation Corpus (SHAC), a multi-institutional corpus of de-identified social history sections annotated for SDOH, including substance use, employment, and living status information. We explore the automatic extraction of SDOH information with SHAC in both standoff and inline annotation formats using GPT-4 in a one-shot prompting setting. We compare GPT-4 extraction performance with a high-performing supervised approach and perform thorough error analyses. Our prompt-based GPT-4 method achieved an overall 0.652 F1 on the SHAC test set, similar to the 7th best-performing system among all teams in the n2c2 challenge with SHAC.
The 2022 n2c2/UW Shared Task on Extracting Social Determinants of Health
Jan 13, 2023Abstract:Objective: The n2c2/UW SDOH Challenge explores the extraction of social determinant of health (SDOH) information from clinical notes. The objectives include the advancement of natural language processing (NLP) information extraction techniques for SDOH and clinical information more broadly. This paper presents the shared task, data, participating teams, performance results, and considerations for future work. Materials and Methods: The task used the Social History Annotated Corpus (SHAC), which consists of clinical text with detailed event-based annotations for SDOH events such as alcohol, drug, tobacco, employment, and living situation. Each SDOH event is characterized through attributes related to status, extent, and temporality. The task includes three subtasks related to information extraction (Subtask A), generalizability (Subtask B), and learning transfer (Subtask C). In addressing this task, participants utilized a range of techniques, including rules, knowledge bases, n-grams, word embeddings, and pretrained language models (LM). Results: A total of 15 teams participated, and the top teams utilized pretrained deep learning LM. The top team across all subtasks used a sequence-to-sequence approach achieving 0.901 F1 for Subtask A, 0.774 F1 Subtask B, and 0.889 F1 for Subtask C. Conclusions: Similar to many NLP tasks and domains, pretrained LM yielded the best performance, including generalizability and learning transfer. An error analysis indicates extraction performance varies by SDOH, with lower performance achieved for conditions, like substance use and homelessness, that increase health risks (risk factors) and higher performance achieved for conditions, like substance abstinence and living with family, that reduce health risks (protective factors).
Extracting Medication Changes in Clinical Narratives using Pre-trained Language Models
Aug 17, 2022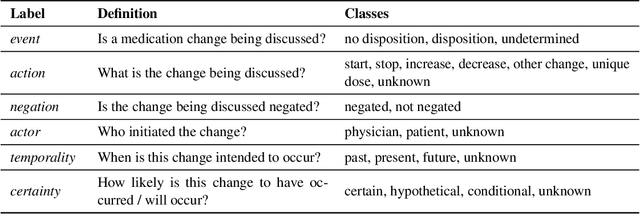
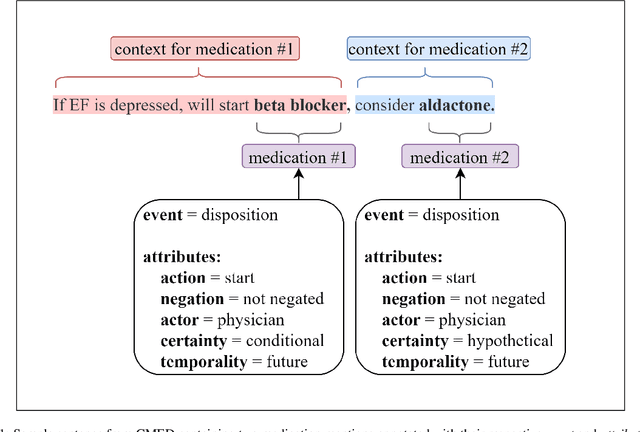
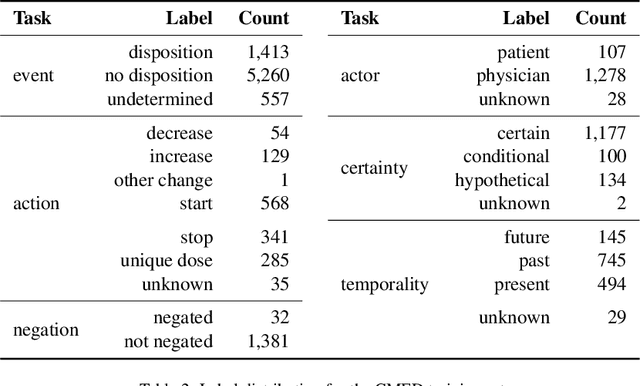
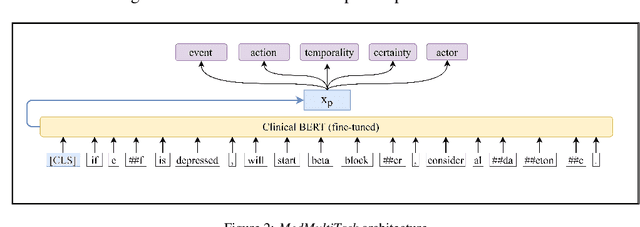
Abstract:An accurate and detailed account of patient medications, including medication changes within the patient timeline, is essential for healthcare providers to provide appropriate patient care. Healthcare providers or the patients themselves may initiate changes to patient medication. Medication changes take many forms, including prescribed medication and associated dosage modification. These changes provide information about the overall health of the patient and the rationale that led to the current care. Future care can then build on the resulting state of the patient. This work explores the automatic extraction of medication change information from free-text clinical notes. The Contextual Medication Event Dataset (CMED) is a corpus of clinical notes with annotations that characterize medication changes through multiple change-related attributes, including the type of change (start, stop, increase, etc.), initiator of the change, temporality, change likelihood, and negation. Using CMED, we identify medication mentions in clinical text and propose three novel high-performing BERT-based systems that resolve the annotated medication change characteristics. We demonstrate that our proposed architectures improve medication change classification performance over the initial work exploring CMED. We identify medication mentions with high performance at 0.959 F1, and our proposed systems classify medication changes and their attributes at an overall average of 0.827 F1.
Performance of Automatic De-identification Across Different Note Types
Feb 17, 2021

Abstract:Free-text clinical notes detail all aspects of patient care and have great potential to facilitate quality improvement and assurance initiatives as well as advance clinical research. However, concerns about patient privacy and confidentiality limit the use of clinical notes for research. As a result, the information documented in these notes remains unavailable for most researchers. De-identification (de-id), i.e., locating and removing personally identifying protected health information (PHI), is one way of improving access to clinical narratives. However, there are limited off-the-shelf de-identification systems able to consistently detect PHI across different data sources and medical specialties. In this abstract, we present the performance of a state-of-the art de-id system called NeuroNER1 on a diverse set of notes from University of Washington (UW) when the models are trained on data from an external institution (Partners Healthcare) vs. from the same institution (UW). We present results at the level of PHI and note types.
Transferability of Neural Network-based De-identification Systems
Feb 17, 2021



Abstract:Methods and Materials: We investigated transferability of neural network-based de-identification sys-tems with and without domain generalization. We used two domain generalization approaches: a novel approach Joint-Domain Learning (JDL) as developed in this paper, and a state-of-the-art domain general-ization approach Common-Specific Decomposition (CSD) from the literature. First, we measured trans-ferability from a single external source. Second, we used two external sources and evaluated whether domain generalization can improve transferability of de-identification models across domains which rep-resent different note types from the same institution. Third, using two external sources with in-domain training data, we studied whether external source data are useful even in cases where sufficient in-domain training data are available. Finally, we investigated transferability of the de-identification mod-els across institutions. Results and Conclusions: We found transferability from a single external source gave inconsistent re-sults. Using additional external sources consistently yielded an F1-score of approximately 80%, but domain generalization was not always helpful to improve transferability. We also found that external sources were useful even in cases where in-domain training data were available by reducing the amount of needed in-domain training data or by improving performance. Transferability across institutions was differed by note type and annotation label. External sources from a different institution were also useful to further improve performance.
A Context-Enhanced De-identification System
Feb 17, 2021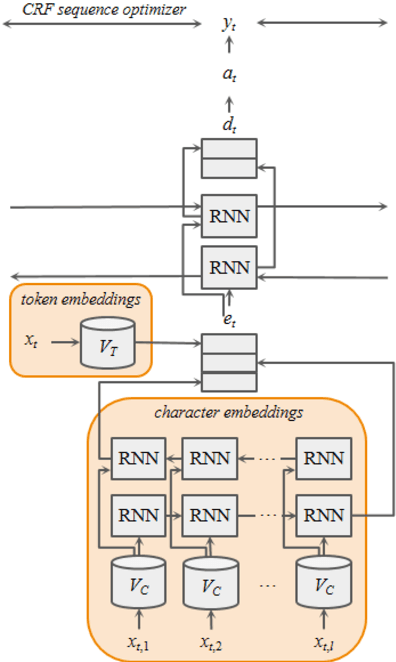
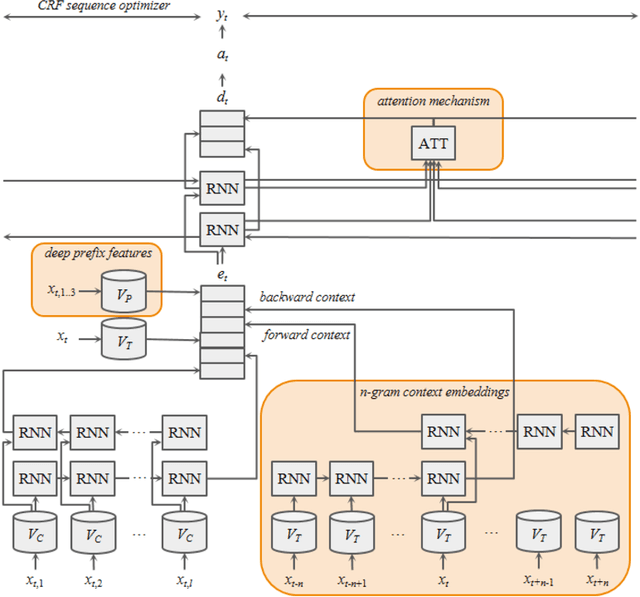
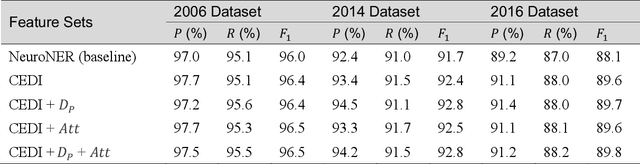
Abstract:Many modern entity recognition systems, including the current state-of-the-art de-identification systems, are based on bidirectional long short-term memory (biLSTM) units augmented by a conditional random field (CRF) sequence optimizer. These systems process the input sentence by sentence. This approach prevents the systems from capturing dependencies over sentence boundaries and makes accurate sentence boundary detection a prerequisite. Since sentence boundary detection can be problematic especially in clinical reports, where dependencies and co-references across sentence boundaries are abundant, these systems have clear limitations. In this study, we built a new system on the framework of one of the current state-of-the-art de-identification systems, NeuroNER, to overcome these limitations. This new system incorporates context embeddings through forward and backward n-grams without using sentence boundaries. Our context-enhanced de-identification (CEDI) system captures dependencies over sentence boundaries and bypasses the sentence boundary detection problem altogether. We enhanced this system with deep affix features and an attention mechanism to capture the pertinent parts of the input. The CEDI system outperforms NeuroNER on the 2006 i2b2 de-identification challenge dataset, the 2014 i2b2 shared task de-identification dataset, and the 2016 CEGS N-GRID de-identification dataset (p<0.01). All datasets comprise narrative clinical reports in English but contain different note types varying from discharge summaries to psychiatric notes. Enhancing CEDI with deep affix features and the attention mechanism further increased performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge