Haritha Gangavarapu
Adapting Biomedical Abstracts into Plain language using Large Language Models
Jan 26, 2025



Abstract:A vast amount of medical knowledge is available for public use through online health forums, and question-answering platforms on social media. The majority of the population in the United States doesn't have the right amount of health literacy to make the best use of that information. Health literacy means the ability to obtain and comprehend the basic health information to make appropriate health decisions. To build the bridge between this gap, organizations advocate adapting this medical knowledge into plain language. Building robust systems to automate the adaptations helps both medical and non-medical professionals best leverage the available information online. The goal of the Plain Language Adaptation of Biomedical Abstracts (PLABA) track is to adapt the biomedical abstracts in English language extracted from PubMed based on the questions asked in MedlinePlus for the general public using plain language at the sentence level. As part of this track, we leveraged the best open-source Large Language Models suitable and fine-tuned for dialog use cases. We compare and present the results for all of our systems and our ranking among the other participants' submissions. Our top performing GPT-4 based model ranked first in the avg. simplicity measure and 3rd on the avg. accuracy measure.
MasonNLP+ at SemEval-2023 Task 8: Extracting Medical Questions, Experiences and Claims from Social Media using Knowledge-Augmented Pre-trained Language Models
Apr 26, 2023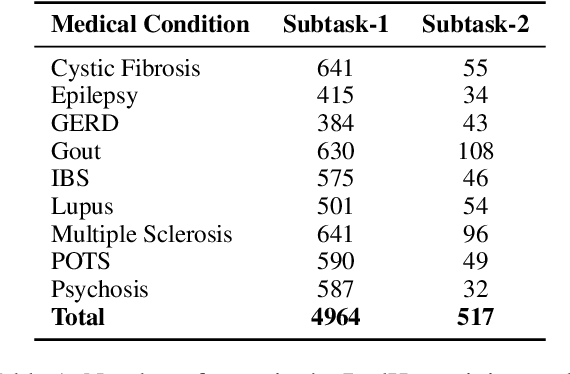



Abstract:In online forums like Reddit, users share their experiences with medical conditions and treatments, including making claims, asking questions, and discussing the effects of treatments on their health. Building systems to understand this information can effectively monitor the spread of misinformation and verify user claims. The Task-8 of the 2023 International Workshop on Semantic Evaluation focused on medical applications, specifically extracting patient experience- and medical condition-related entities from user posts on social media. The Reddit Health Online Talk (RedHot) corpus contains posts from medical condition-related subreddits with annotations characterizing the patient experience and medical conditions. In Subtask-1, patient experience is characterized by personal experience, questions, and claims. In Subtask-2, medical conditions are characterized by population, intervention, and outcome. For the automatic extraction of patient experiences and medical condition information, as a part of the challenge, we proposed language-model-based extraction systems that ranked $3^{rd}$ on both subtasks' leaderboards. In this work, we describe our approach and, in addition, explore the automatic extraction of this information using domain-specific language models and the inclusion of external knowledge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge