Nicholas J Dobbins
Generalizable and Scalable Multistage Biomedical Concept Normalization Leveraging Large Language Models
May 24, 2024Abstract:Background: Biomedical entity normalization is critical to biomedical research because the richness of free-text clinical data, such as progress notes, can often be fully leveraged only after translating words and phrases into structured and coded representations suitable for analysis. Large Language Models (LLMs), in turn, have shown great potential and high performance in a variety of natural language processing (NLP) tasks, but their application for normalization remains understudied. Methods: We applied both proprietary and open-source LLMs in combination with several rule-based normalization systems commonly used in biomedical research. We used a two-step LLM integration approach, (1) using an LLM to generate alternative phrasings of a source utterance, and (2) to prune candidate UMLS concepts, using a variety of prompting methods. We measure results by $F_{\beta}$, where we favor recall over precision, and F1. Results: We evaluated a total of 5,523 concept terms and text contexts from a publicly available dataset of human-annotated biomedical abstracts. Incorporating GPT-3.5-turbo increased overall $F_{\beta}$ and F1 in normalization systems +9.5 and +7.3 (MetaMapLite), +13.9 and +10.9 (QuickUMLS), and +10.5 and +10.3 (BM25), while the open-source Vicuna model achieved +10.8 and +12.2 (MetaMapLite), +14.7 and +15 (QuickUMLS), and +15.6 and +18.7 (BM25). Conclusions: Existing general-purpose LLMs, both propriety and open-source, can be leveraged at scale to greatly improve normalization performance using existing tools, with no fine-tuning.
Extracting Social Determinants of Health from Pediatric Patient Notes Using Large Language Models: Novel Corpus and Methods
Apr 04, 2024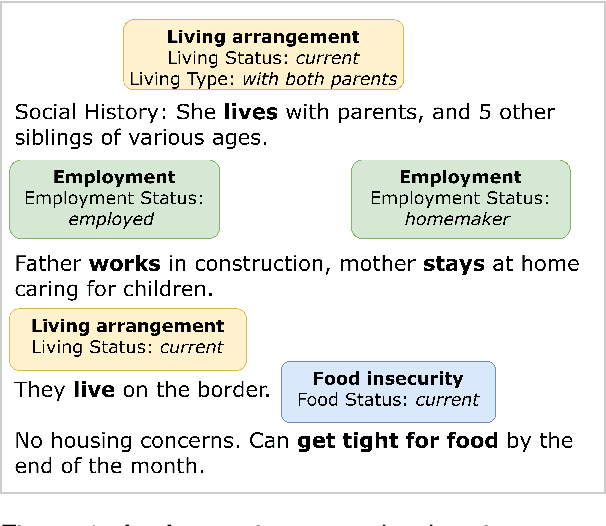
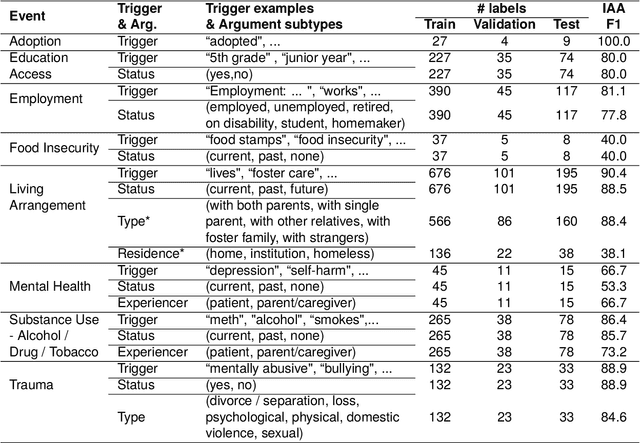
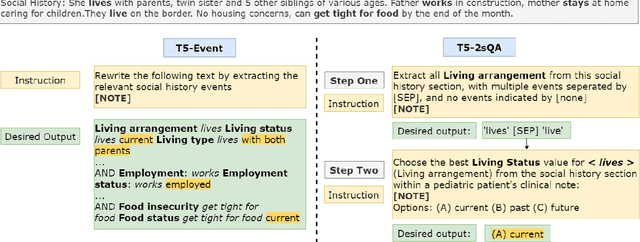
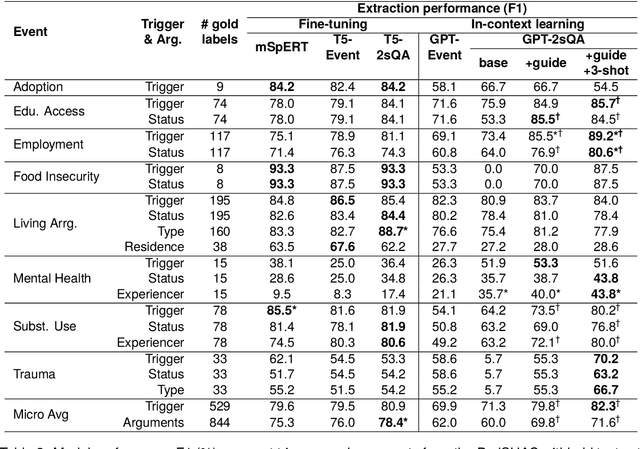
Abstract:Social determinants of health (SDoH) play a critical role in shaping health outcomes, particularly in pediatric populations where interventions can have long-term implications. SDoH are frequently studied in the Electronic Health Record (EHR), which provides a rich repository for diverse patient data. In this work, we present a novel annotated corpus, the Pediatric Social History Annotation Corpus (PedSHAC), and evaluate the automatic extraction of detailed SDoH representations using fine-tuned and in-context learning methods with Large Language Models (LLMs). PedSHAC comprises annotated social history sections from 1,260 clinical notes obtained from pediatric patients within the University of Washington (UW) hospital system. Employing an event-based annotation scheme, PedSHAC captures ten distinct health determinants to encompass living and economic stability, prior trauma, education access, substance use history, and mental health with an overall annotator agreement of 81.9 F1. Our proposed fine-tuning LLM-based extractors achieve high performance at 78.4 F1 for event arguments. In-context learning approaches with GPT-4 demonstrate promise for reliable SDoH extraction with limited annotated examples, with extraction performance at 82.3 F1 for event triggers.
Prompt-based Extraction of Social Determinants of Health Using Few-shot Learning
Jun 12, 2023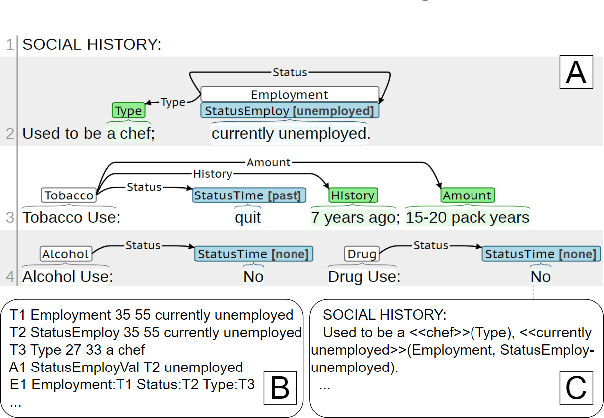
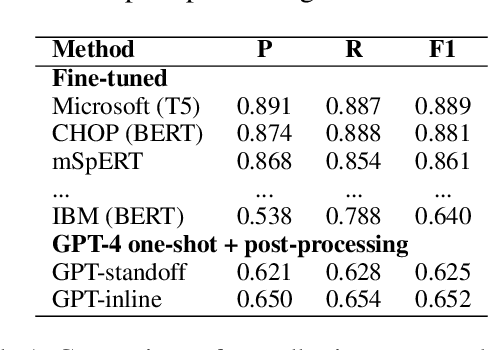
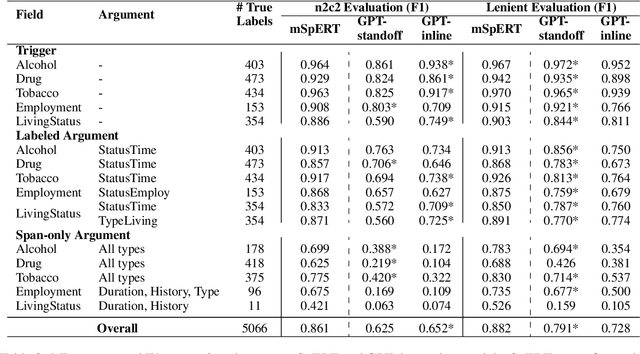
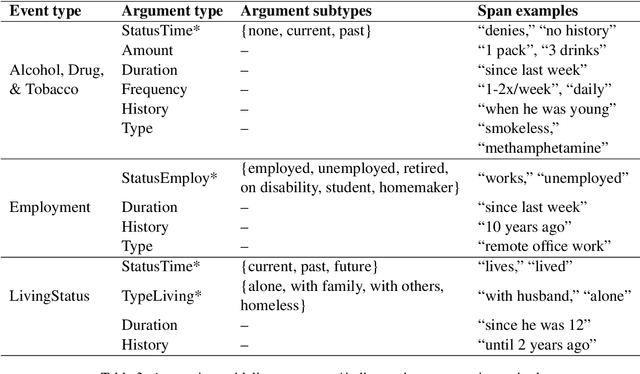
Abstract:Social determinants of health (SDOH) documented in the electronic health record through unstructured text are increasingly being studied to understand how SDOH impacts patient health outcomes. In this work, we utilize the Social History Annotation Corpus (SHAC), a multi-institutional corpus of de-identified social history sections annotated for SDOH, including substance use, employment, and living status information. We explore the automatic extraction of SDOH information with SHAC in both standoff and inline annotation formats using GPT-4 in a one-shot prompting setting. We compare GPT-4 extraction performance with a high-performing supervised approach and perform thorough error analyses. Our prompt-based GPT-4 method achieved an overall 0.652 F1 on the SHAC test set, similar to the 7th best-performing system among all teams in the n2c2 challenge with SHAC.
LeafAI: query generator for clinical cohort discovery rivaling a human programmer
Apr 13, 2023



Abstract:Objective: Identifying study-eligible patients within clinical databases is a critical step in clinical research. However, accurate query design typically requires extensive technical and biomedical expertise. We sought to create a system capable of generating data model-agnostic queries while also providing novel logical reasoning capabilities for complex clinical trial eligibility criteria. Materials and Methods: The task of query creation from eligibility criteria requires solving several text-processing problems, including named entity recognition and relation extraction, sequence-to-sequence transformation, normalization, and reasoning. We incorporated hybrid deep learning and rule-based modules for these, as well as a knowledge base of the Unified Medical Language System (UMLS) and linked ontologies. To enable data-model agnostic query creation, we introduce a novel method for tagging database schema elements using UMLS concepts. To evaluate our system, called LeafAI, we compared the capability of LeafAI to a human database programmer to identify patients who had been enrolled in 8 clinical trials conducted at our institution. We measured performance by the number of actual enrolled patients matched by generated queries. Results: LeafAI matched a mean 43% of enrolled patients with 27,225 eligible across 8 clinical trials, compared to 27% matched and 14,587 eligible in queries by a human database programmer. The human programmer spent 26 total hours crafting queries compared to several minutes by LeafAI. Conclusions: Our work contributes a state-of-the-art data model-agnostic query generation system capable of conditional reasoning using a knowledge base. We demonstrate that LeafAI can rival a human programmer in finding patients eligible for clinical trials.
Leveraging Natural Language Processing to Augment Structured Social Determinants of Health Data in the Electronic Health Record
Dec 14, 2022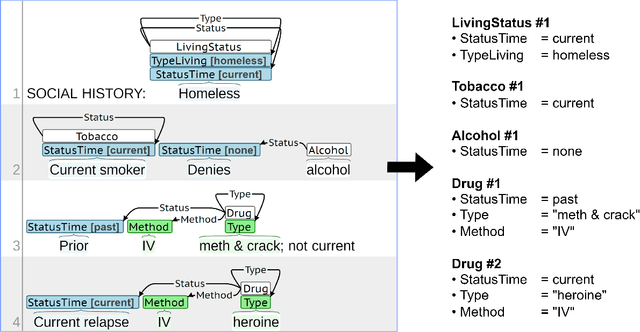


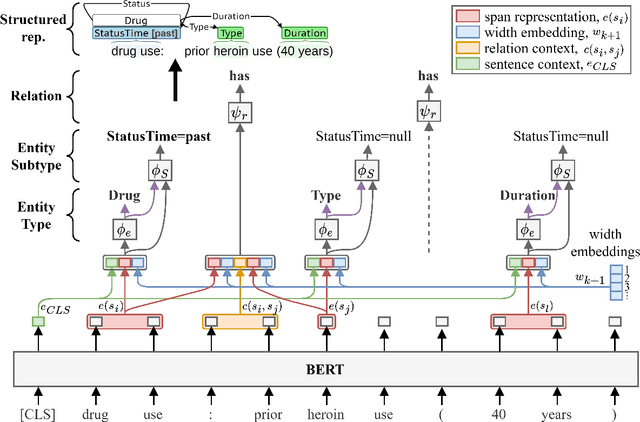
Abstract:Objective: Social Determinants of Health (SDOH) influence personal health outcomes and health systems interactions. Health systems capture SDOH information through structured data and unstructured clinical notes; however, clinical notes often contain a more comprehensive representation of several key SDOH. The objective of this work is to assess the SDOH information gain achievable by extracting structured semantic representations of SDOH from the clinical narrative and combining these extracted representations with available structured data. Materials and Methods: We developed a natural language processing (NLP) information extraction model for SDOH that utilizes a deep learning entity and relation extraction architecture. In an electronic health record (EHR) case study, we applied the SDOH extractor to a large existing clinical data set with over 200,000 patients and 400,000 notes and compared the extracted information with available structured data. Results: The SDOH extractor achieved 0.86 F1 on a withheld test set. In the EHR case study, we found 19\% of current tobacco users, 10\% of drug users, and 32\% of homeless patients only include documentation of these risk factors in the clinical narrative. Conclusions: Patients who are at-risk for negative health outcomes due to SDOH may be better served if health systems are able to identify SDOH risk factors and associated social needs. Structured semantic representations of text-encoded SDOH information can augment existing structured, and this more comprehensive SDOH representation can assist health systems in identifying and addressing social needs.
The Leaf Clinical Trials Corpus: a new resource for query generation from clinical trial eligibility criteria
Jul 27, 2022Abstract:Identifying cohorts of patients based on eligibility criteria such as medical conditions, procedures, and medication use is critical to recruitment for clinical trials. Such criteria are often most naturally described in free-text, using language familiar to clinicians and researchers. In order to identify potential participants at scale, these criteria must first be translated into queries on clinical databases, which can be labor-intensive and error-prone. Natural language processing (NLP) methods offer a potential means of such conversion into database queries automatically. However they must first be trained and evaluated using corpora which capture clinical trials criteria in sufficient detail. In this paper, we introduce the Leaf Clinical Trials (LCT) corpus, a human-annotated corpus of over 1,000 clinical trial eligibility criteria descriptions using highly granular structured labels capturing a range of biomedical phenomena. We provide details of our schema, annotation process, corpus quality, and statistics. Additionally, we present baseline information extraction results on this corpus as benchmarks for future work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge