Namu Park
Identifying Imaging Follow-Up in Radiology Reports: A Comparative Analysis of Traditional ML and LLM Approaches
Nov 14, 2025Abstract:Large language models (LLMs) have shown considerable promise in clinical natural language processing, yet few domain-specific datasets exist to rigorously evaluate their performance on radiology tasks. In this work, we introduce an annotated corpus of 6,393 radiology reports from 586 patients, each labeled for follow-up imaging status, to support the development and benchmarking of follow-up adherence detection systems. Using this corpus, we systematically compared traditional machine-learning classifiers, including logistic regression (LR), support vector machines (SVM), Longformer, and a fully fine-tuned Llama3-8B-Instruct, with recent generative LLMs. To evaluate generative LLMs, we tested GPT-4o and the open-source GPT-OSS-20B under two configurations: a baseline (Base) and a task-optimized (Advanced) setting that focused inputs on metadata, recommendation sentences, and their surrounding context. A refined prompt for GPT-OSS-20B further improved reasoning accuracy. Performance was assessed using precision, recall, and F1 scores with 95% confidence intervals estimated via non-parametric bootstrapping. Inter-annotator agreement was high (F1 = 0.846). GPT-4o (Advanced) achieved the best performance (F1 = 0.832), followed closely by GPT-OSS-20B (Advanced; F1 = 0.828). LR and SVM also performed strongly (F1 = 0.776 and 0.775), underscoring that while LLMs approach human-level agreement through prompt optimization, interpretable and resource-efficient models remain valuable baselines.
BioMistral-NLU: Towards More Generalizable Medical Language Understanding through Instruction Tuning
Oct 24, 2024



Abstract:Large language models (LLMs) such as ChatGPT are fine-tuned on large and diverse instruction-following corpora, and can generalize to new tasks. However, those instruction-tuned LLMs often perform poorly in specialized medical natural language understanding (NLU) tasks that require domain knowledge, granular text comprehension, and structured data extraction. To bridge the gap, we: (1) propose a unified prompting format for 7 important NLU tasks, % through span extraction and multi-choice question-answering (QA), (2) curate an instruction-tuning dataset, MNLU-Instruct, utilizing diverse existing open-source medical NLU corpora, and (3) develop BioMistral-NLU, a generalizable medical NLU model, through fine-tuning BioMistral on MNLU-Instruct. We evaluate BioMistral-NLU in a zero-shot setting, across 6 important NLU tasks, from two widely adopted medical NLU benchmarks: Biomedical Language Understanding Evaluation (BLUE) and Biomedical Language Understanding and Reasoning Benchmark (BLURB). Our experiments show that our BioMistral-NLU outperforms the original BioMistral, as well as the proprietary LLMs - ChatGPT and GPT-4. Our dataset-agnostic prompting strategy and instruction tuning step over diverse NLU tasks enhance LLMs' generalizability across diverse medical NLU tasks. Our ablation experiments show that instruction-tuning on a wider variety of tasks, even when the total number of training instances remains constant, enhances downstream zero-shot generalization.
Extracting Social Determinants of Health from Pediatric Patient Notes Using Large Language Models: Novel Corpus and Methods
Apr 04, 2024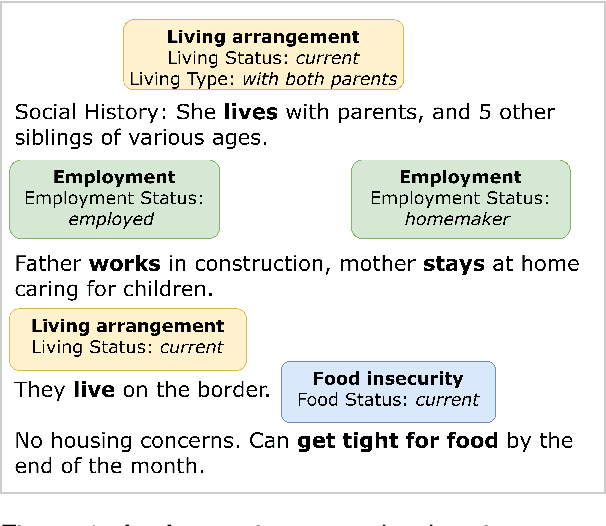
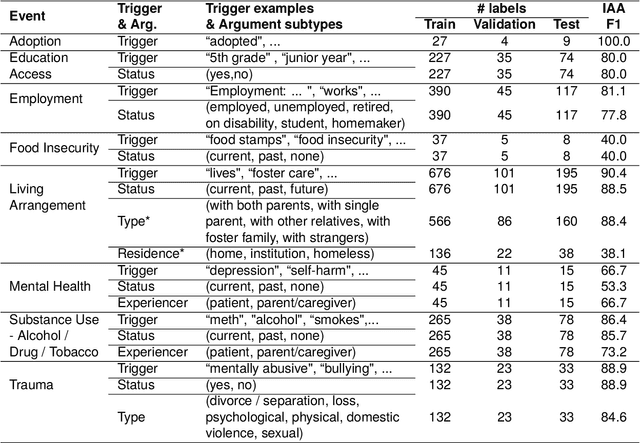
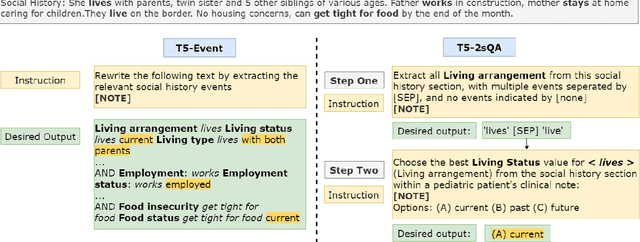
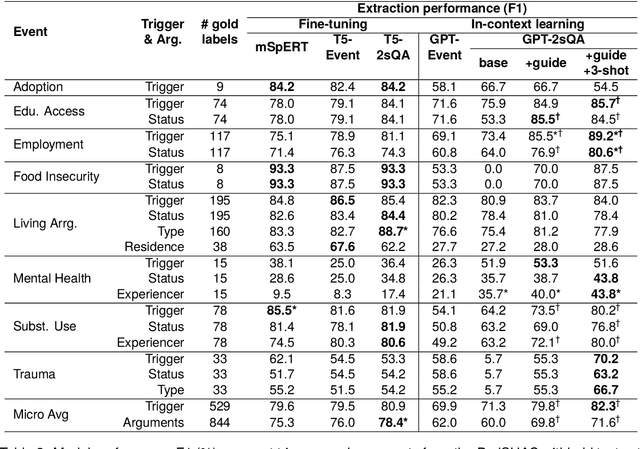
Abstract:Social determinants of health (SDoH) play a critical role in shaping health outcomes, particularly in pediatric populations where interventions can have long-term implications. SDoH are frequently studied in the Electronic Health Record (EHR), which provides a rich repository for diverse patient data. In this work, we present a novel annotated corpus, the Pediatric Social History Annotation Corpus (PedSHAC), and evaluate the automatic extraction of detailed SDoH representations using fine-tuned and in-context learning methods with Large Language Models (LLMs). PedSHAC comprises annotated social history sections from 1,260 clinical notes obtained from pediatric patients within the University of Washington (UW) hospital system. Employing an event-based annotation scheme, PedSHAC captures ten distinct health determinants to encompass living and economic stability, prior trauma, education access, substance use history, and mental health with an overall annotator agreement of 81.9 F1. Our proposed fine-tuning LLM-based extractors achieve high performance at 78.4 F1 for event arguments. In-context learning approaches with GPT-4 demonstrate promise for reliable SDoH extraction with limited annotated examples, with extraction performance at 82.3 F1 for event triggers.
A Novel Corpus of Annotated Medical Imaging Reports and Information Extraction Results Using BERT-based Language Models
Mar 27, 2024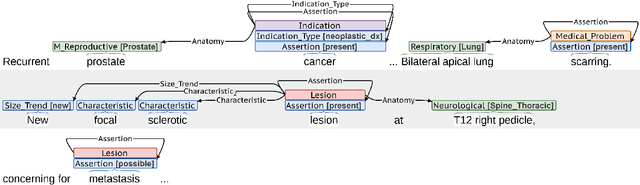
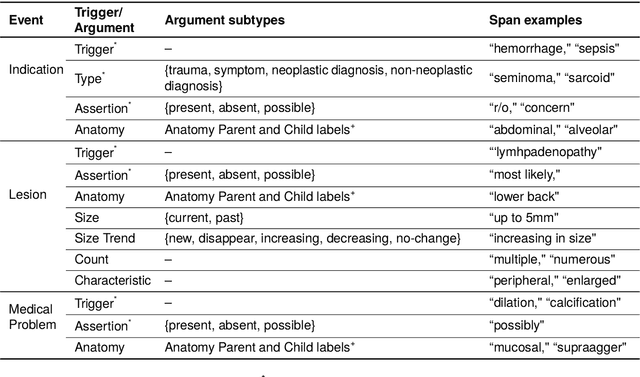
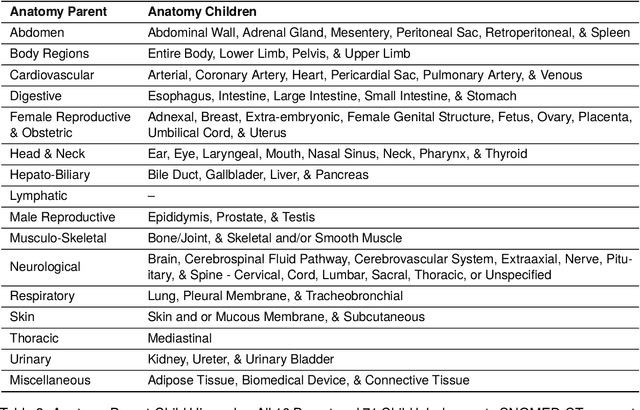
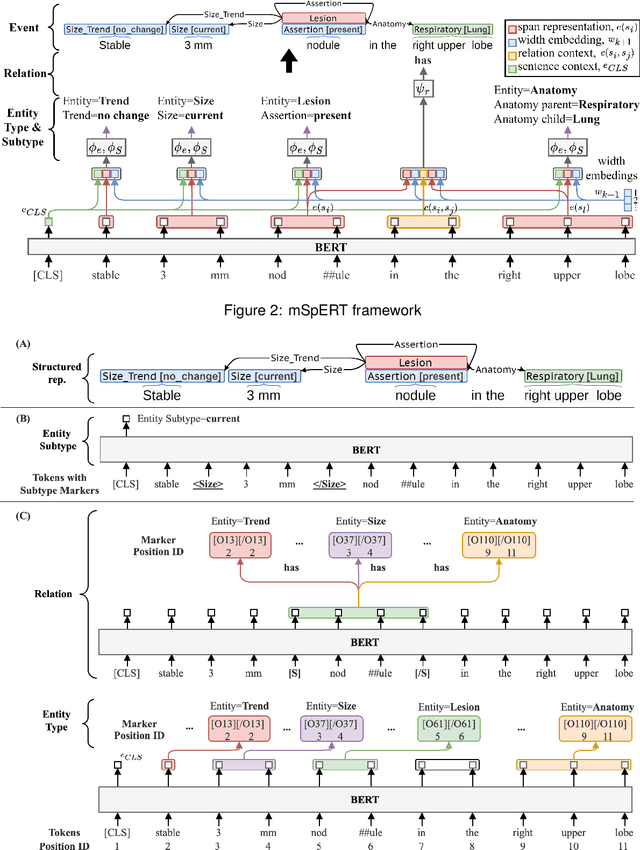
Abstract:Medical imaging is critical to the diagnosis, surveillance, and treatment of many health conditions, including oncological, neurological, cardiovascular, and musculoskeletal disorders, among others. Radiologists interpret these complex, unstructured images and articulate their assessments through narrative reports that remain largely unstructured. This unstructured narrative must be converted into a structured semantic representation to facilitate secondary applications such as retrospective analyses or clinical decision support. Here, we introduce the Corpus of Annotated Medical Imaging Reports (CAMIR), which includes 609 annotated radiology reports from three imaging modality types: Computed Tomography, Magnetic Resonance Imaging, and Positron Emission Tomography-Computed Tomography. Reports were annotated using an event-based schema that captures clinical indications, lesions, and medical problems. Each event consists of a trigger and multiple arguments, and a majority of the argument types, including anatomy, normalize the spans to pre-defined concepts to facilitate secondary use. CAMIR uniquely combines a granular event structure and concept normalization. To extract CAMIR events, we explored two BERT (Bi-directional Encoder Representation from Transformers)-based architectures, including an existing architecture (mSpERT) that jointly extracts all event information and a multi-step approach (PL-Marker++) that we augmented for the CAMIR schema.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge