Weipeng Zhou
A Federated and Parameter-Efficient Framework for Large Language Model Training in Medicine
Jan 29, 2026Abstract:Large language models (LLMs) have demonstrated strong performance on medical benchmarks, including question answering and diagnosis. To enable their use in clinical settings, LLMs are typically further adapted through continued pretraining or post-training using clinical data. However, most medical LLMs are trained on data from a single institution, which faces limitations in generalizability and safety in heterogeneous systems. Federated learning (FL) is a promising solution for enabling collaborative model development across healthcare institutions. Yet applying FL to LLMs in medicine remains fundamentally limited. First, conventional FL requires transmitting the full model during each communication round, which becomes impractical for multi-billion-parameter LLMs given the limited computational resources. Second, many FL algorithms implicitly assume data homogeneity, whereas real-world clinical data are highly heterogeneous across patients, diseases, and institutional practices. We introduce the model-agnostic and parameter-efficient federated learning framework for adapting LLMs to medical applications. Fed-MedLoRA transmits only low-rank adapter parameters, reducing communication and computation overhead, while Fed-MedLoRA+ further incorporates adaptive, data-aware aggregation to improve convergence under cross-site heterogeneity. We apply the framework to clinical information extraction (IE), which transforms patient narratives into structured medical entities and relations. Accuracy was assessed across five patient cohorts through comparisons with BERT models, and LLaMA-3 and DeepSeek-R1, GPT-4o models. Evaluation settings included (1) in-domain training and testing, (2) external validation on independent cohorts, and (3) a low-resource new-site adaptation scenario using real-world clinical notes from the Yale New Haven Health System.
Fino1: On the Transferability of Reasoning Enhanced LLMs to Finance
Feb 12, 2025Abstract:Recent advancements in large language models (LLMs) have shown strong general reasoning abilities, yet their effectiveness in financial reasoning remains underexplored. In this study, we comprehensively evaluate 16 powerful reasoning and general LLMs on three complex financial tasks involving financial text, tabular data, and equations, assessing numerical reasoning, tabular interpretation, financial terminology comprehension, long-context processing, and equation-based problem solving. Our results show that while better datasets and pretraining improve financial reasoning, general enhancements like CoT fine-tuning do not always yield consistent gains. Moreover, all reasoning strategies face challenges in improving performance on long-context and multi-table tasks. To address these limitations, we develop a financial reasoning-enhanced model based on Llama-3.1-8B-Instruct, by CoT fine-tuning and reinforcement learning with domain-specific reasoning paths. Even with simple fine-tuning with one financial dataset, our model achieves a consistent 10% performance improvement across tasks, surpassing all 8B models and even Llama3-70B-Instruct and Llama3.1-70B-Instruct on average. Our results highlight the need for domain-specific adaptations in financial tasks, emphasizing future directions such as multi-table reasoning, long-context processing, and financial terminology comprehension. All our datasets, models, and codes are publicly available. Furthermore, we introduce a leaderboard for benchmarking future datasets and models.
LeafAI: query generator for clinical cohort discovery rivaling a human programmer
Apr 13, 2023



Abstract:Objective: Identifying study-eligible patients within clinical databases is a critical step in clinical research. However, accurate query design typically requires extensive technical and biomedical expertise. We sought to create a system capable of generating data model-agnostic queries while also providing novel logical reasoning capabilities for complex clinical trial eligibility criteria. Materials and Methods: The task of query creation from eligibility criteria requires solving several text-processing problems, including named entity recognition and relation extraction, sequence-to-sequence transformation, normalization, and reasoning. We incorporated hybrid deep learning and rule-based modules for these, as well as a knowledge base of the Unified Medical Language System (UMLS) and linked ontologies. To enable data-model agnostic query creation, we introduce a novel method for tagging database schema elements using UMLS concepts. To evaluate our system, called LeafAI, we compared the capability of LeafAI to a human database programmer to identify patients who had been enrolled in 8 clinical trials conducted at our institution. We measured performance by the number of actual enrolled patients matched by generated queries. Results: LeafAI matched a mean 43% of enrolled patients with 27,225 eligible across 8 clinical trials, compared to 27% matched and 14,587 eligible in queries by a human database programmer. The human programmer spent 26 total hours crafting queries compared to several minutes by LeafAI. Conclusions: Our work contributes a state-of-the-art data model-agnostic query generation system capable of conditional reasoning using a knowledge base. We demonstrate that LeafAI can rival a human programmer in finding patients eligible for clinical trials.
Forecasting Daily COVID-19 Related Calls in VA Health Care System: Predictive Model Development
Nov 30, 2021

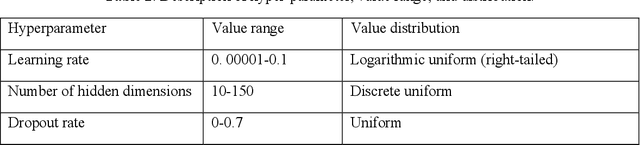
Abstract:Background: COVID-19 has become a challenge worldwide and properly planning of medical resources is the key to combating COVID-19. In the US Veteran Affairs Health Care System (VA), many of the enrollees are susceptible to COVID-19. Predicting the COVID-19 to allocate medical resources promptly becomes a critical issue. When the VA enrollees have COVID-19 symptoms, it is recommended that their first step should be to call the VA Call Center. For confirmed COVID-19 patients, the median time from the first symptom to hospital admission was seven days. By predicting the number of COVID-19 related calls, we could predict imminent surges in healthcare use and plan medical resources ahead. Objective: The study aims to develop a method to forecast the daily number of COVID-19 related calls for each of the 110 VA medical centers. Methods: In the proposed method, we pre-trained a model using a cluster of medical centers and fine-tuned it for individual medical centers. At the cluster level, we performed feature selection to select significant features and automatic hyper-parameter search to select optimal hyper-parameter value combinations for the model. Conclusions: This study proposed an accurate method to forecast the daily number of COVID-19 related calls for VA medical centers. The proposed method was able to overcome modeling challenges by grouping similar medical centers into clusters to enlarge the dataset for training models, and using hyper-parameter search to automatically find optimal hyper-parameter value combinations for models. With the proposed method, surges in health care can be predicted ahead. This allows health care practitioners to better plan medical resources and combat COVID-19.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge