Bridget T. McInnes
CACER: Clinical Concept Annotations for Cancer Events and Relations
Sep 05, 2024
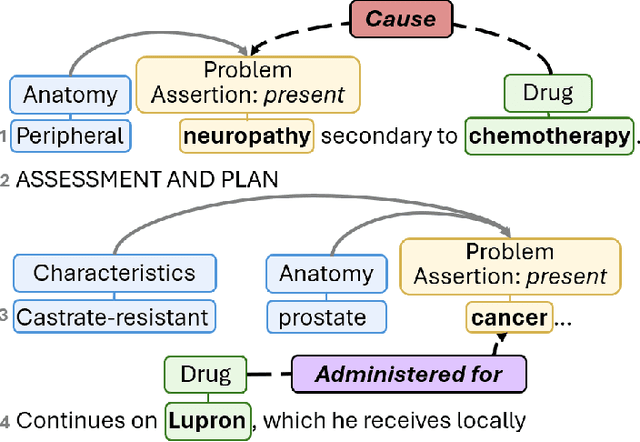
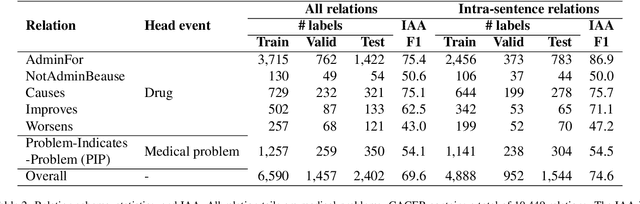
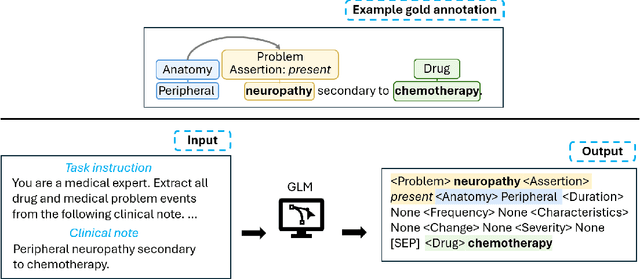
Abstract:Clinical notes contain unstructured representations of patient histories, including the relationships between medical problems and prescription drugs. To investigate the relationship between cancer drugs and their associated symptom burden, we extract structured, semantic representations of medical problem and drug information from the clinical narratives of oncology notes. We present Clinical Concept Annotations for Cancer Events and Relations (CACER), a novel corpus with fine-grained annotations for over 48,000 medical problems and drug events and 10,000 drug-problem and problem-problem relations. Leveraging CACER, we develop and evaluate transformer-based information extraction (IE) models such as BERT, Flan-T5, Llama3, and GPT-4 using fine-tuning and in-context learning (ICL). In event extraction, the fine-tuned BERT and Llama3 models achieved the highest performance at 88.2-88.0 F1, which is comparable to the inter-annotator agreement (IAA) of 88.4 F1. In relation extraction, the fine-tuned BERT, Flan-T5, and Llama3 achieved the highest performance at 61.8-65.3 F1. GPT-4 with ICL achieved the worst performance across both tasks. The fine-tuned models significantly outperformed GPT-4 in ICL, highlighting the importance of annotated training data and model optimization. Furthermore, the BERT models performed similarly to Llama3. For our task, LLMs offer no performance advantage over the smaller BERT models. The results emphasize the need for annotated training data to optimize models. Multiple fine-tuned transformer models achieved performance comparable to IAA for several extraction tasks.
* This is a pre-copy-editing, author-produced PDF of an article accepted for publication in JAMIA following peer review. The definitive publisher-authenticated version is available online at https://academic.oup.com/jamia/advance-article/doi/10.1093/jamia/ocae231/7748302
BioBERT-based Deep Learning and Merged ChemProt-DrugProt for Enhanced Biomedical Relation Extraction
May 28, 2024Abstract:This paper presents a methodology for enhancing relation extraction from biomedical texts, focusing specifically on chemical-gene interactions. Leveraging the BioBERT model and a multi-layer fully connected network architecture, our approach integrates the ChemProt and DrugProt datasets using a novel merging strategy. Through extensive experimentation, we demonstrate significant performance improvements, particularly in CPR groups shared between the datasets. The findings underscore the importance of dataset merging in augmenting sample counts and improving model accuracy. Moreover, the study highlights the potential of automated information extraction in biomedical research and clinical practice.
Extracting Adverse Drug Events from Clinical Notes
Apr 21, 2021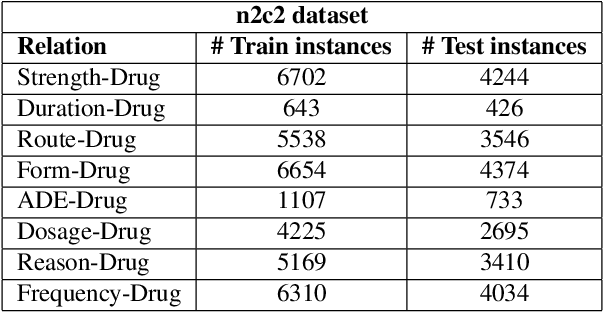
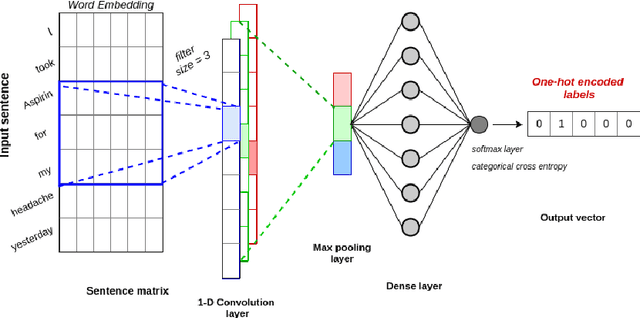
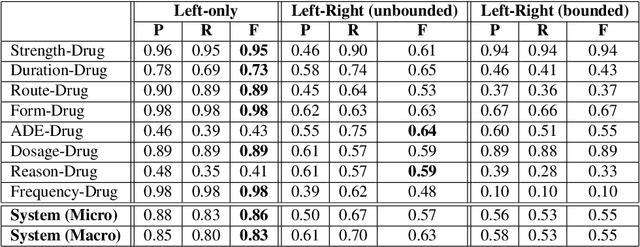
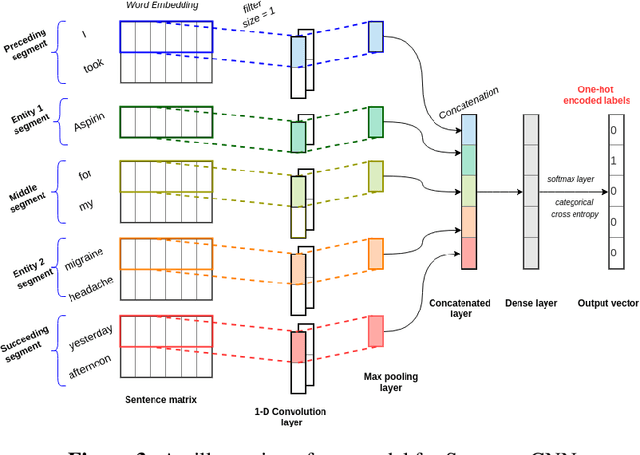
Abstract:Adverse drug events (ADEs) are unexpected incidents caused by the administration of a drug or medication. To identify and extract these events, we require information about not just the drug itself but attributes describing the drug (e.g., strength, dosage), the reason why the drug was initially prescribed, and any adverse reaction to the drug. This paper explores the relationship between a drug and its associated attributes using relation extraction techniques. We explore three approaches: a rule-based approach, a deep learning-based approach, and a contextualized language model-based approach. We evaluate our system on the n2c2-2018 ADE extraction dataset. Our experimental results demonstrate that the contextualized language model-based approach outperformed other models overall and obtain the state-of-the-art performance in ADE extraction with a Precision of 0.93, Recall of 0.96, and an $F_1$ score of 0.94; however, for certain relation types, the rule-based approach obtained a higher Precision and Recall than either learning approach.
* 9 pages, 4 figures
MT-Clinical BERT: Scaling Clinical Information Extraction with Multitask Learning
Apr 21, 2020



Abstract:Clinical notes contain an abundance of important but not-readily accessible information about patients. Systems to automatically extract this information rely on large amounts of training data for which their exists limited resources to create. Furthermore, they are developed dis-jointly; meaning that no information can be shared amongst task-specific systems. This bottle-neck unnecessarily complicates practical application, reduces the performance capabilities of each individual solution and associates the engineering debt of managing multiple information extraction systems. We address these challenges by developing Multitask-Clinical BERT: a single deep learning model that simultaneously performs eight clinical tasks spanning entity extraction, PHI identification, language entailment and similarity by sharing representations amongst tasks. We find our single system performs competitively with all state-the-art task-specific systems while also benefiting from massive computational benefits at inference.
Improving Correlation with Human Judgments by Integrating Semantic Similarity with Second--Order Vectors
May 27, 2017



Abstract:Vector space methods that measure semantic similarity and relatedness often rely on distributional information such as co--occurrence frequencies or statistical measures of association to weight the importance of particular co--occurrences. In this paper, we extend these methods by incorporating a measure of semantic similarity based on a human curated taxonomy into a second--order vector representation. This results in a measure of semantic relatedness that combines both the contextual information available in a corpus--based vector space representation with the semantic knowledge found in a biomedical ontology. Our results show that incorporating semantic similarity into a second order co--occurrence matrices improves correlation with human judgments for both similarity and relatedness, and that our method compares favorably to various different word embedding methods that have recently been evaluated on the same reference standards we have used.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge