Darshini Mahendran
BioBERT-based Deep Learning and Merged ChemProt-DrugProt for Enhanced Biomedical Relation Extraction
May 28, 2024Abstract:This paper presents a methodology for enhancing relation extraction from biomedical texts, focusing specifically on chemical-gene interactions. Leveraging the BioBERT model and a multi-layer fully connected network architecture, our approach integrates the ChemProt and DrugProt datasets using a novel merging strategy. Through extensive experimentation, we demonstrate significant performance improvements, particularly in CPR groups shared between the datasets. The findings underscore the importance of dataset merging in augmenting sample counts and improving model accuracy. Moreover, the study highlights the potential of automated information extraction in biomedical research and clinical practice.
Extracting Adverse Drug Events from Clinical Notes
Apr 21, 2021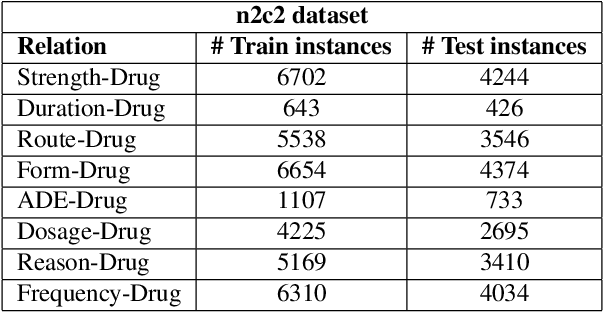
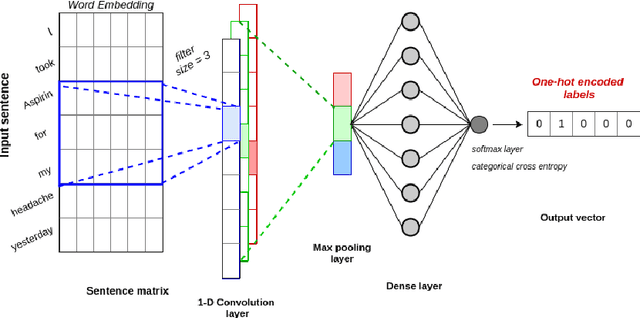
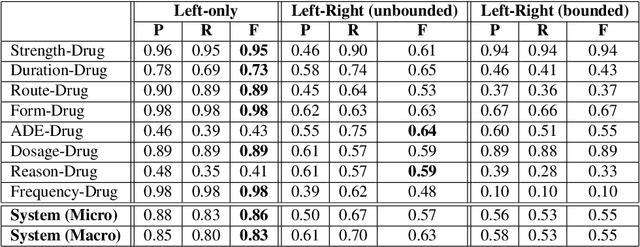
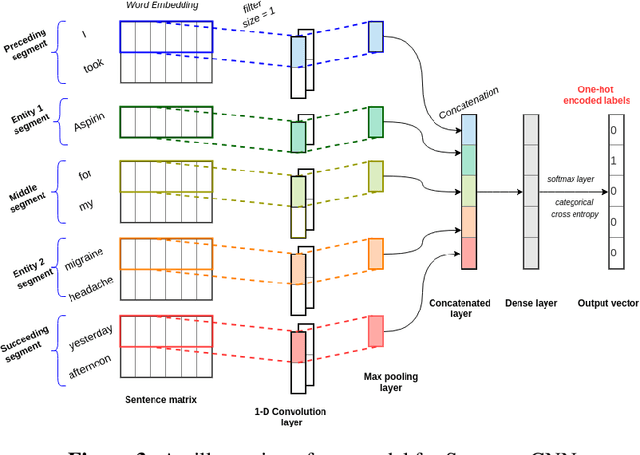
Abstract:Adverse drug events (ADEs) are unexpected incidents caused by the administration of a drug or medication. To identify and extract these events, we require information about not just the drug itself but attributes describing the drug (e.g., strength, dosage), the reason why the drug was initially prescribed, and any adverse reaction to the drug. This paper explores the relationship between a drug and its associated attributes using relation extraction techniques. We explore three approaches: a rule-based approach, a deep learning-based approach, and a contextualized language model-based approach. We evaluate our system on the n2c2-2018 ADE extraction dataset. Our experimental results demonstrate that the contextualized language model-based approach outperformed other models overall and obtain the state-of-the-art performance in ADE extraction with a Precision of 0.93, Recall of 0.96, and an $F_1$ score of 0.94; however, for certain relation types, the rule-based approach obtained a higher Precision and Recall than either learning approach.
* 9 pages, 4 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge