Diwakar Mahajan
BioVERSE: Representation Alignment of Biomedical Modalities to LLMs for Multi-Modal Reasoning
Oct 01, 2025Abstract:Recent advances in large language models (LLMs) and biomedical foundation models (BioFMs) have achieved strong results in biological text reasoning, molecular modeling, and single-cell analysis, yet they remain siloed in disjoint embedding spaces, limiting cross-modal reasoning. We present BIOVERSE (Biomedical Vector Embedding Realignment for Semantic Engagement), a two-stage approach that adapts pretrained BioFMs as modality encoders and aligns them with LLMs through lightweight, modality-specific projection layers. The approach first aligns each modality to a shared LLM space through independently trained projections, allowing them to interoperate naturally, and then applies standard instruction tuning with multi-modal data to bring them together for downstream reasoning. By unifying raw biomedical data with knowledge embedded in LLMs, the approach enables zero-shot annotation, cross-modal question answering, and interactive, explainable dialogue. Across tasks spanning cell-type annotation, molecular description, and protein function reasoning, compact BIOVERSE configurations surpass larger LLM baselines while enabling richer, generative outputs than existing BioFMs, establishing a foundation for principled multi-modal biomedical reasoning.
Multi-view biomedical foundation models for molecule-target and property prediction
Oct 25, 2024


Abstract:Foundation models applied to bio-molecular space hold promise to accelerate drug discovery. Molecular representation is key to building such models. Previous works have typically focused on a single representation or view of the molecules. Here, we develop a multi-view foundation model approach, that integrates molecular views of graph, image and text. Single-view foundation models are each pre-trained on a dataset of up to 200M molecules and then aggregated into combined representations. Our multi-view model is validated on a diverse set of 18 tasks, encompassing ligand-protein binding, molecular solubility, metabolism and toxicity. We show that the multi-view models perform robustly and are able to balance the strengths and weaknesses of specific views. We then apply this model to screen compounds against a large (>100 targets) set of G Protein-Coupled receptors (GPCRs). From this library of targets, we identify 33 that are related to Alzheimer's disease. On this subset, we employ our model to identify strong binders, which are validated through structure-based modeling and identification of key binding motifs.
MISMATCH: Fine-grained Evaluation of Machine-generated Text with Mismatch Error Types
Jun 18, 2023


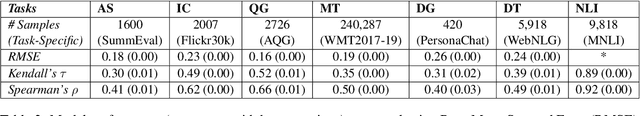
Abstract:With the growing interest in large language models, the need for evaluating the quality of machine text compared to reference (typically human-generated) text has become focal attention. Most recent works focus either on task-specific evaluation metrics or study the properties of machine-generated text captured by the existing metrics. In this work, we propose a new evaluation scheme to model human judgments in 7 NLP tasks, based on the fine-grained mismatches between a pair of texts. Inspired by the recent efforts in several NLP tasks for fine-grained evaluation, we introduce a set of 13 mismatch error types such as spatial/geographic errors, entity errors, etc, to guide the model for better prediction of human judgments. We propose a neural framework for evaluating machine texts that uses these mismatch error types as auxiliary tasks and re-purposes the existing single-number evaluation metrics as additional scalar features, in addition to textual features extracted from the machine and reference texts. Our experiments reveal key insights about the existing metrics via the mismatch errors. We show that the mismatch errors between the sentence pairs on the held-out datasets from 7 NLP tasks align well with the human evaluation.
Do We Still Need Clinical Language Models?
Feb 16, 2023



Abstract:Although recent advances in scaling large language models (LLMs) have resulted in improvements on many NLP tasks, it remains unclear whether these models trained primarily with general web text are the right tool in highly specialized, safety critical domains such as clinical text. Recent results have suggested that LLMs encode a surprising amount of medical knowledge. This raises an important question regarding the utility of smaller domain-specific language models. With the success of general-domain LLMs, is there still a need for specialized clinical models? To investigate this question, we conduct an extensive empirical analysis of 12 language models, ranging from 220M to 175B parameters, measuring their performance on 3 different clinical tasks that test their ability to parse and reason over electronic health records. As part of our experiments, we train T5-Base and T5-Large models from scratch on clinical notes from MIMIC III and IV to directly investigate the efficiency of clinical tokens. We show that relatively small specialized clinical models substantially outperform all in-context learning approaches, even when finetuned on limited annotated data. Further, we find that pretraining on clinical tokens allows for smaller, more parameter-efficient models that either match or outperform much larger language models trained on general text. We release the code and the models used under the PhysioNet Credentialed Health Data license and data use agreement.
Extracting Medication Changes in Clinical Narratives using Pre-trained Language Models
Aug 17, 2022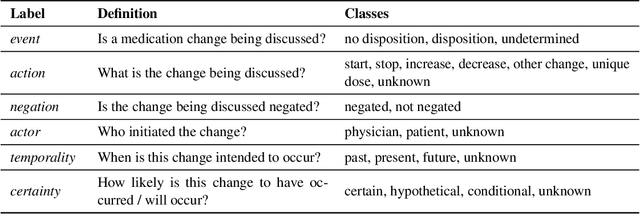
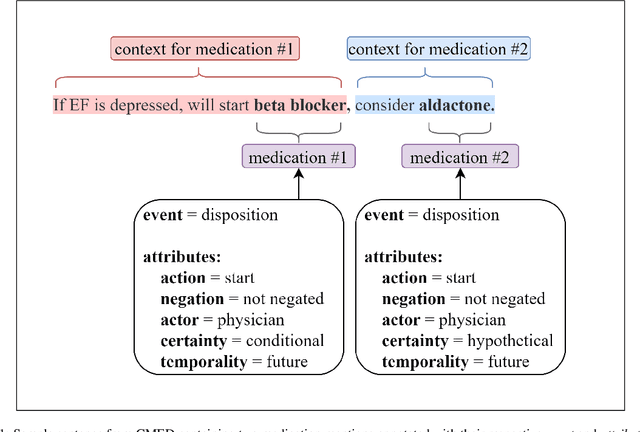
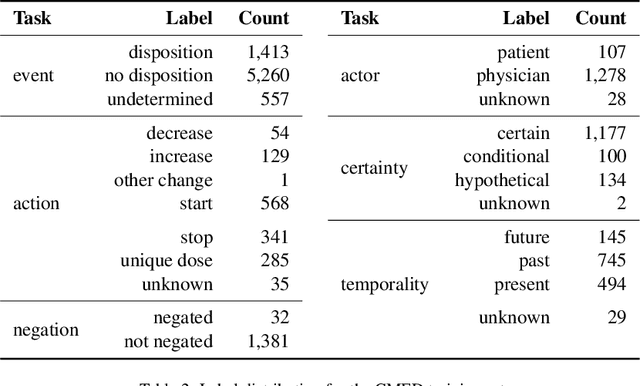
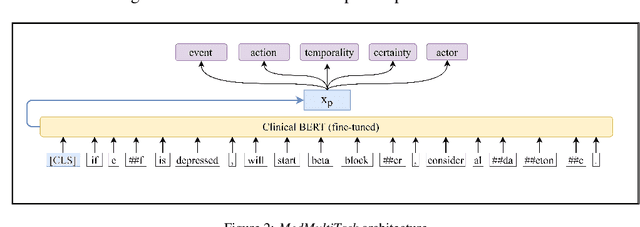
Abstract:An accurate and detailed account of patient medications, including medication changes within the patient timeline, is essential for healthcare providers to provide appropriate patient care. Healthcare providers or the patients themselves may initiate changes to patient medication. Medication changes take many forms, including prescribed medication and associated dosage modification. These changes provide information about the overall health of the patient and the rationale that led to the current care. Future care can then build on the resulting state of the patient. This work explores the automatic extraction of medication change information from free-text clinical notes. The Contextual Medication Event Dataset (CMED) is a corpus of clinical notes with annotations that characterize medication changes through multiple change-related attributes, including the type of change (start, stop, increase, etc.), initiator of the change, temporality, change likelihood, and negation. Using CMED, we identify medication mentions in clinical text and propose three novel high-performing BERT-based systems that resolve the annotated medication change characteristics. We demonstrate that our proposed architectures improve medication change classification performance over the initial work exploring CMED. We identify medication mentions with high performance at 0.959 F1, and our proposed systems classify medication changes and their attributes at an overall average of 0.827 F1.
SemEval-2021 Task 9: Fact Verification and Evidence Finding for Tabular Data in Scientific Documents
May 28, 2021



Abstract:Understanding tables is an important and relevant task that involves understanding table structure as well as being able to compare and contrast information within cells. In this paper, we address this challenge by presenting a new dataset and tasks that addresses this goal in a shared task in SemEval 2020 Task 9: Fact Verification and Evidence Finding for Tabular Data in Scientific Documents (SEM-TAB-FACTS). Our dataset contains 981 manually-generated tables and an auto-generated dataset of 1980 tables providing over 180K statement and over 16M evidence annotations. SEM-TAB-FACTS featured two sub-tasks. In sub-task A, the goal was to determine if a statement is supported, refuted or unknown in relation to a table. In sub-task B, the focus was on identifying the specific cells of a table that provide evidence for the statement. 69 teams signed up to participate in the task with 19 successful submissions to subtask A and 12 successful submissions to subtask B. We present our results and main findings from the competition.
Toward Understanding Clinical Context of Medication Change Events in Clinical Narratives
Nov 17, 2020



Abstract:Understanding medication events in clinical narratives is essential to achieving a complete picture of a patient's medication history. While prior research has explored identification of medication changes in clinical notes, due to the longitudinal and narrative nature of clinical documentation, extraction of medication change alone without the necessary clinical context is insufficient for use in real-world applications, such as medication timeline generation and medication reconciliation. In this paper, we present the Contextualized Medication Event Dataset (CMED), a dataset for capturing relevant context of medication changes documented in clinical notes, which was developed using a novel conceptual framework that organizes context for clinical events into various orthogonal dimensions. In this process, we define specific contextual aspects pertinent to medication change events (i.e. Action, Negation, Temporality, Certainty, and Actor), describe the annotation process and challenges encountered, and report the results of preliminary experiments. The resulting dataset, CMED, consists of 9,013 medication mentions annotated over 500 clinical notes. To encourage development of methods for improved understanding of medications in clinical narratives, CMED will be released to the community as a shared task in 2021.
WNTRAC: Artificial Intelligence Assisted Tracking of Non-pharmaceutical Interventions Implemented Worldwide for COVID-19
Sep 16, 2020



Abstract:The Coronavirus disease 2019 (COVID-19) global pandemic has transformed almost every facet of human society throughout the world. Against an emerging, highly transmissible disease with no definitive treatment or vaccine, governments worldwide have implemented non-pharmaceutical intervention (NPI) to slow the spread of the virus. Examples of such interventions include community actions (e.g. school closures, restrictions on mass gatherings), individual actions (e.g. mask wearing, self-quarantine), and environmental actions (e.g. public facility cleaning). We present the Worldwide Non-pharmaceutical Interventions Tracker for COVID-19 (WNTRAC), a comprehensive dataset consisting of over 6,000 NPIs implemented worldwide since the start of the pandemic. WNTRAC covers NPIs implemented across 261 countries and territories, and classifies NPI measures into a taxonomy of sixteen NPI types. NPI measures are automatically extracted daily from Wikipedia articles using natural language processing techniques and manually validated to ensure accuracy and veracity. We hope that the dataset is valuable for policymakers, public health leaders, and researchers in modeling and analysis efforts for controlling the spread of COVID-19.
Extracting Daily Dosage from Medication Instructions in EHRs: An Automated Approach and Lessons Learned
May 21, 2020
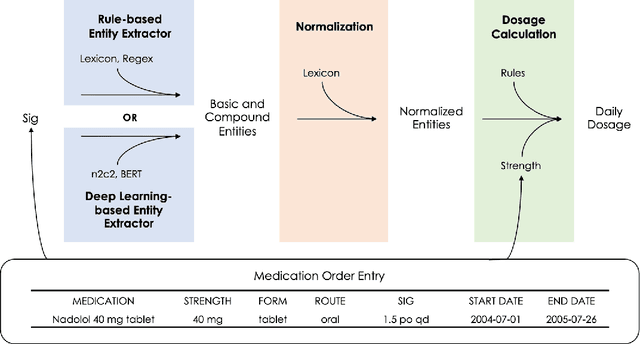

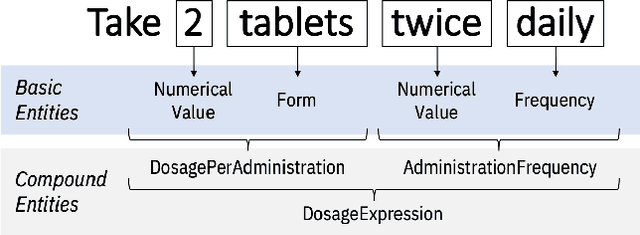
Abstract:Understanding a patient's medication history is essential for physicians to provide appropriate treatment recommendations. A medication's prescribed daily dosage is a key element of the medication history; however, it is generally not provided as a discrete quantity and needs to be derived from free text medication instructions (Sigs) in the structured electronic health record (EHR). Existing works in daily dosage extraction are narrow in scope, dealing with dosage extraction for a single drug from clinical notes. Here, we present an automated approach to calculate daily dosage for all medications in EHR structured data. We describe and characterize the variable language used in Sigs, and present our hybrid system for calculating daily dosage combining deep learning-based named entity extractor with lexicon dictionaries and regular expressions. Our system achieves 0.98 precision and 0.95 recall on an expert-generated dataset of 1000 Sigs, demonstrating its effectiveness on the general purpose daily dosage calculation task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge