Daniel E. Platt
Multi-view biomedical foundation models for molecule-target and property prediction
Oct 25, 2024


Abstract:Foundation models applied to bio-molecular space hold promise to accelerate drug discovery. Molecular representation is key to building such models. Previous works have typically focused on a single representation or view of the molecules. Here, we develop a multi-view foundation model approach, that integrates molecular views of graph, image and text. Single-view foundation models are each pre-trained on a dataset of up to 200M molecules and then aggregated into combined representations. Our multi-view model is validated on a diverse set of 18 tasks, encompassing ligand-protein binding, molecular solubility, metabolism and toxicity. We show that the multi-view models perform robustly and are able to balance the strengths and weaknesses of specific views. We then apply this model to screen compounds against a large (>100 targets) set of G Protein-Coupled receptors (GPCRs). From this library of targets, we identify 33 that are related to Alzheimer's disease. On this subset, we employ our model to identify strong binders, which are validated through structure-based modeling and identification of key binding motifs.
Inferring COVID-19 Biological Pathways from Clinical Phenotypes via Topological Analysis
Jan 19, 2021
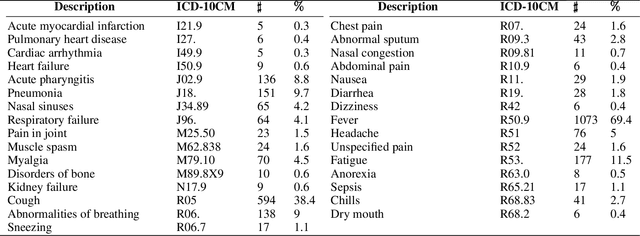
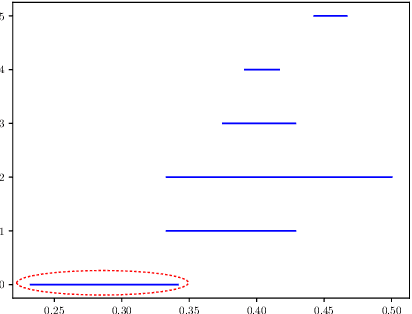
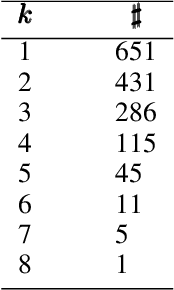
Abstract:COVID-19 has caused thousands of deaths around the world and also resulted in a large international economic disruption. Identifying the pathways associated with this illness can help medical researchers to better understand the properties of the condition. This process can be carried out by analyzing the medical records. It is crucial to develop tools and models that can aid researchers with this process in a timely manner. However, medical records are often unstructured clinical notes, and this poses significant challenges to developing the automated systems. In this article, we propose a pipeline to aid practitioners in analyzing clinical notes and revealing the pathways associated with this disease. Our pipeline relies on topological properties and consists of three steps: 1) pre-processing the clinical notes to extract the salient concepts, 2) constructing a feature space of the patients to characterize the extracted concepts, and finally, 3) leveraging the topological properties to distill the available knowledge and visualize the result. Our experiments on a publicly available dataset of COVID-19 clinical notes testify that our pipeline can indeed extract meaningful pathways.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge