Eric Lehman
BioClinical ModernBERT: A State-of-the-Art Long-Context Encoder for Biomedical and Clinical NLP
Jun 12, 2025Abstract:Encoder-based transformer models are central to biomedical and clinical Natural Language Processing (NLP), as their bidirectional self-attention makes them well-suited for efficiently extracting structured information from unstructured text through discriminative tasks. However, encoders have seen slower development compared to decoder models, leading to limited domain adaptation in biomedical and clinical settings. We introduce BioClinical ModernBERT, a domain-adapted encoder that builds on the recent ModernBERT release, incorporating long-context processing and substantial improvements in speed and performance for biomedical and clinical NLP. BioClinical ModernBERT is developed through continued pretraining on the largest biomedical and clinical corpus to date, with over 53.5 billion tokens, and addresses a key limitation of prior clinical encoders by leveraging 20 datasets from diverse institutions, domains, and geographic regions, rather than relying on data from a single source. It outperforms existing biomedical and clinical encoders on four downstream tasks spanning a broad range of use cases. We release both base (150M parameters) and large (396M parameters) versions of BioClinical ModernBERT, along with training checkpoints to support further research.
From Sparse to Dense: GPT-4 Summarization with Chain of Density Prompting
Sep 08, 2023Abstract:Selecting the ``right'' amount of information to include in a summary is a difficult task. A good summary should be detailed and entity-centric without being overly dense and hard to follow. To better understand this tradeoff, we solicit increasingly dense GPT-4 summaries with what we refer to as a ``Chain of Density'' (CoD) prompt. Specifically, GPT-4 generates an initial entity-sparse summary before iteratively incorporating missing salient entities without increasing the length. Summaries generated by CoD are more abstractive, exhibit more fusion, and have less of a lead bias than GPT-4 summaries generated by a vanilla prompt. We conduct a human preference study on 100 CNN DailyMail articles and find that that humans prefer GPT-4 summaries that are more dense than those generated by a vanilla prompt and almost as dense as human written summaries. Qualitative analysis supports the notion that there exists a tradeoff between informativeness and readability. 500 annotated CoD summaries, as well as an extra 5,000 unannotated summaries, are freely available on HuggingFace (https://huggingface.co/datasets/griffin/chain_of_density).
Do We Still Need Clinical Language Models?
Feb 16, 2023



Abstract:Although recent advances in scaling large language models (LLMs) have resulted in improvements on many NLP tasks, it remains unclear whether these models trained primarily with general web text are the right tool in highly specialized, safety critical domains such as clinical text. Recent results have suggested that LLMs encode a surprising amount of medical knowledge. This raises an important question regarding the utility of smaller domain-specific language models. With the success of general-domain LLMs, is there still a need for specialized clinical models? To investigate this question, we conduct an extensive empirical analysis of 12 language models, ranging from 220M to 175B parameters, measuring their performance on 3 different clinical tasks that test their ability to parse and reason over electronic health records. As part of our experiments, we train T5-Base and T5-Large models from scratch on clinical notes from MIMIC III and IV to directly investigate the efficiency of clinical tokens. We show that relatively small specialized clinical models substantially outperform all in-context learning approaches, even when finetuned on limited annotated data. Further, we find that pretraining on clinical tokens allows for smaller, more parameter-efficient models that either match or outperform much larger language models trained on general text. We release the code and the models used under the PhysioNet Credentialed Health Data license and data use agreement.
Learning to Ask Like a Physician
Jun 06, 2022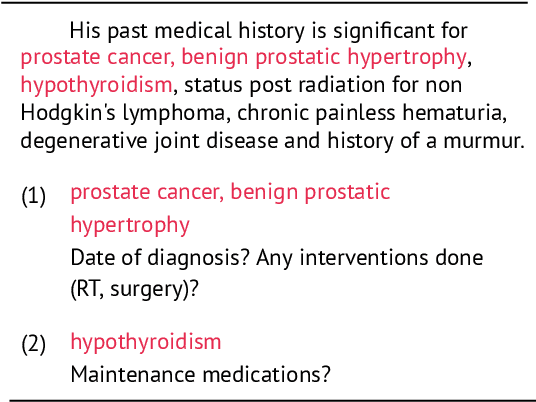
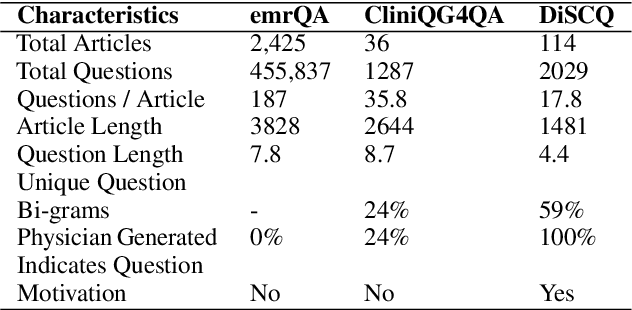
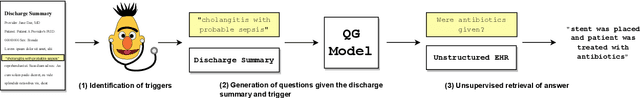
Abstract:Existing question answering (QA) datasets derived from electronic health records (EHR) are artificially generated and consequently fail to capture realistic physician information needs. We present Discharge Summary Clinical Questions (DiSCQ), a newly curated question dataset composed of 2,000+ questions paired with the snippets of text (triggers) that prompted each question. The questions are generated by medical experts from 100+ MIMIC-III discharge summaries. We analyze this dataset to characterize the types of information sought by medical experts. We also train baseline models for trigger detection and question generation (QG), paired with unsupervised answer retrieval over EHRs. Our baseline model is able to generate high quality questions in over 62% of cases when prompted with human selected triggers. We release this dataset (and all code to reproduce baseline model results) to facilitate further research into realistic clinical QA and QG: https://github.com/elehman16/discq.
Does BERT Pretrained on Clinical Notes Reveal Sensitive Data?
Apr 22, 2021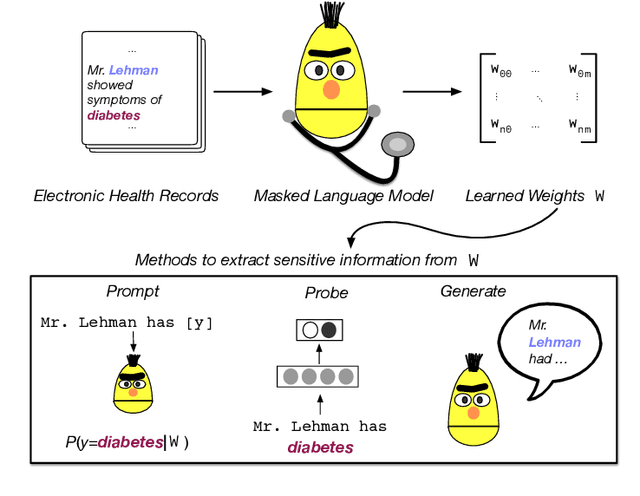

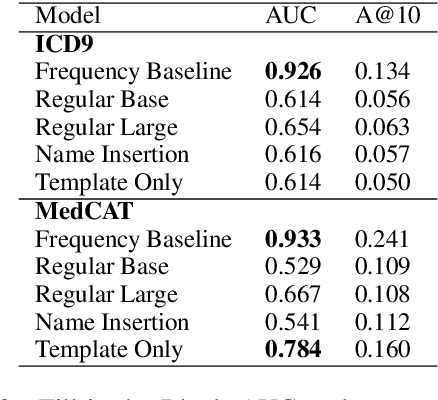
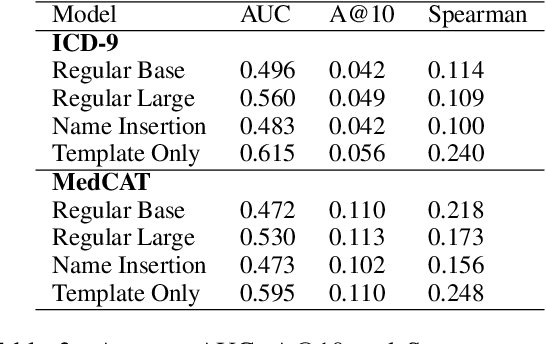
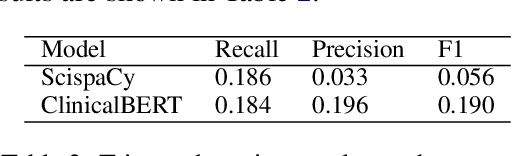
Abstract:Large Transformers pretrained over clinical notes from Electronic Health Records (EHR) have afforded substantial gains in performance on predictive clinical tasks. The cost of training such models (and the necessity of data access to do so) coupled with their utility motivates parameter sharing, i.e., the release of pretrained models such as ClinicalBERT. While most efforts have used deidentified EHR, many researchers have access to large sets of sensitive, non-deidentified EHR with which they might train a BERT model (or similar). Would it be safe to release the weights of such a model if they did? In this work, we design a battery of approaches intended to recover Personal Health Information (PHI) from a trained BERT. Specifically, we attempt to recover patient names and conditions with which they are associated. We find that simple probing methods are not able to meaningfully extract sensitive information from BERT trained over the MIMIC-III corpus of EHR. However, more sophisticated "attacks" may succeed in doing so: To facilitate such research, we make our experimental setup and baseline probing models available at https://github.com/elehman16/exposing_patient_data_release
Understanding Clinical Trial Reports: Extracting Medical Entities and Their Relations
Oct 08, 2020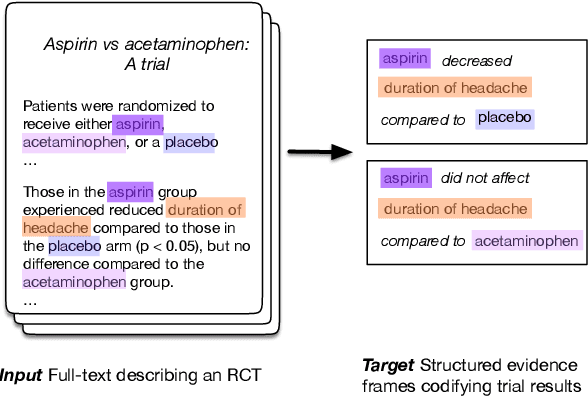
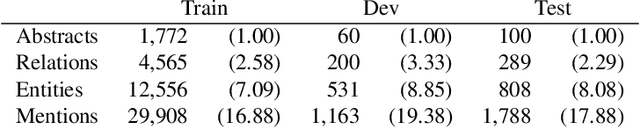
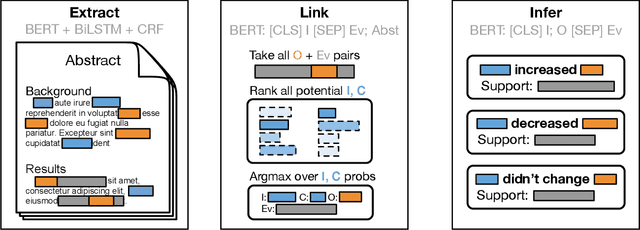

Abstract:The best evidence concerning comparative treatment effectiveness comes from clinical trials, the results of which are reported in unstructured articles. Medical experts must manually extract information from articles to inform decision-making, which is time-consuming and expensive. Here we consider the end-to-end task of both (a) extracting treatments and outcomes from full-text articles describing clinical trials (entity identification) and, (b) inferring the reported results for the former with respect to the latter (relation extraction). We introduce new data for this task, and evaluate models that have recently achieved state-of-the-art results on similar tasks in Natural Language Processing. We then propose a new method motivated by how trial results are typically presented that outperforms these purely data-driven baselines. Finally, we run a fielded evaluation of the model with a non-profit seeking to identify existing drugs that might be re-purposed for cancer, showing the potential utility of end-to-end evidence extraction systems.
Evidence Inference 2.0: More Data, Better Models
May 14, 2020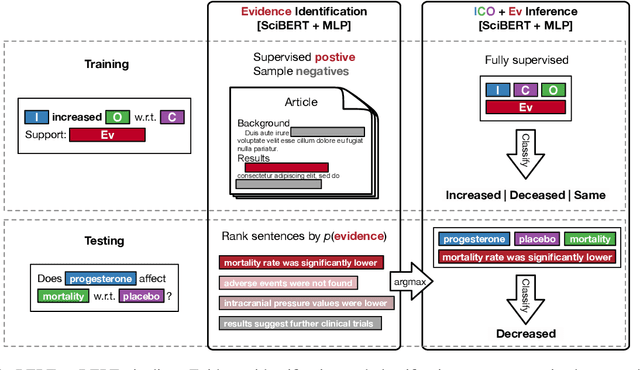


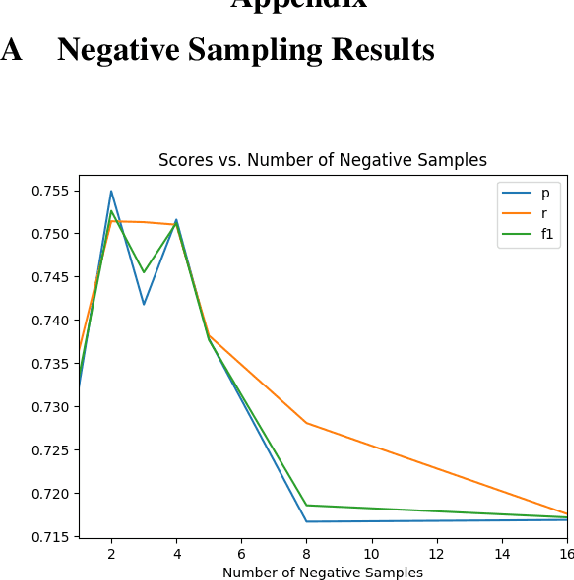
Abstract:How do we most effectively treat a disease or condition? Ideally, we could consult a database of evidence gleaned from clinical trials to answer such questions. Unfortunately, no such database exists; clinical trial results are instead disseminated primarily via lengthy natural language articles. Perusing all such articles would be prohibitively time-consuming for healthcare practitioners; they instead tend to depend on manually compiled systematic reviews of medical literature to inform care. NLP may speed this process up, and eventually facilitate immediate consult of published evidence. The Evidence Inference dataset was recently released to facilitate research toward this end. This task entails inferring the comparative performance of two treatments, with respect to a given outcome, from a particular article (describing a clinical trial) and identifying supporting evidence. For instance: Does this article report that chemotherapy performed better than surgery for five-year survival rates of operable cancers? In this paper, we collect additional annotations to expand the Evidence Inference dataset by 25\%, provide stronger baseline models, systematically inspect the errors that these make, and probe dataset quality. We also release an abstract only (as opposed to full-texts) version of the task for rapid model prototyping. The updated corpus, documentation, and code for new baselines and evaluations are available at http://evidence-inference.ebm-nlp.com/.
ERASER: A Benchmark to Evaluate Rationalized NLP Models
Nov 08, 2019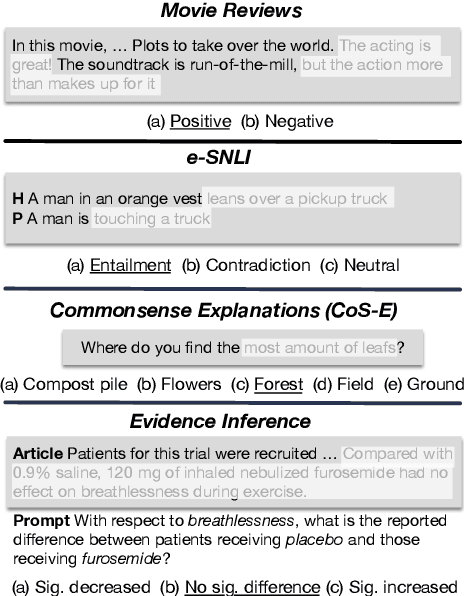
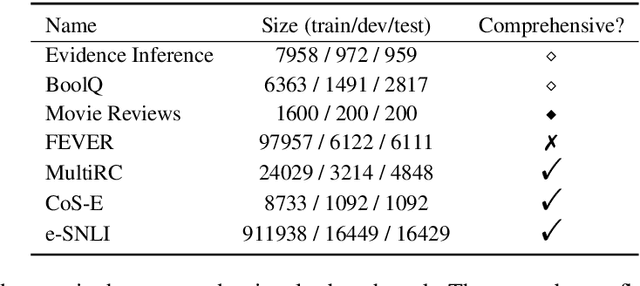
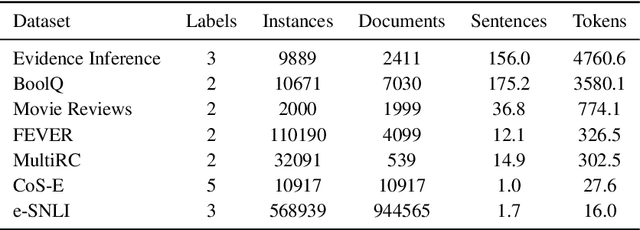
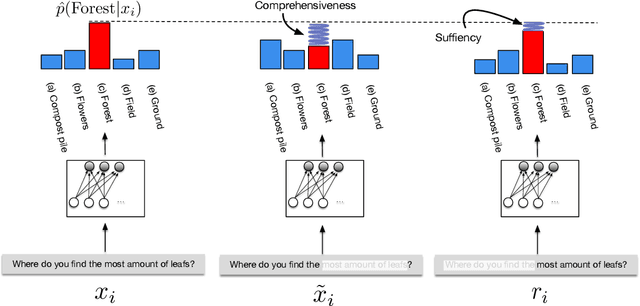
Abstract:State-of-the-art models in NLP are now predominantly based on deep neural networks that are generally opaque in terms of how they come to specific predictions. This limitation has led to increased interest in designing more interpretable deep models for NLP that can reveal the `reasoning' underlying model outputs. But work in this direction has been conducted on different datasets and tasks with correspondingly unique aims and metrics; this makes it difficult to track progress. We propose the Evaluating Rationales And Simple English Reasoning (ERASER) benchmark to advance research on interpretable models in NLP. This benchmark comprises multiple datasets and tasks for which human annotations of "rationales" (supporting evidence) have been collected. We propose several metrics that aim to capture how well the rationales provided by models align with human rationales, and also how faithful these rationales are (i.e., the degree to which provided rationales influenced the corresponding predictions). Our hope is that releasing this benchmark facilitates progress on designing more interpretable NLP systems. The benchmark, code, and documentation are available at: www.eraserbenchmark.com .
Inferring Which Medical Treatments Work from Reports of Clinical Trials
Apr 04, 2019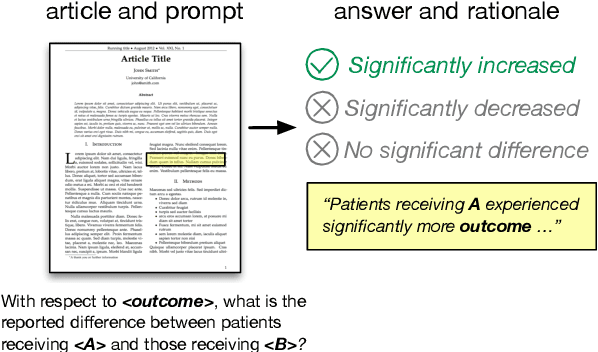

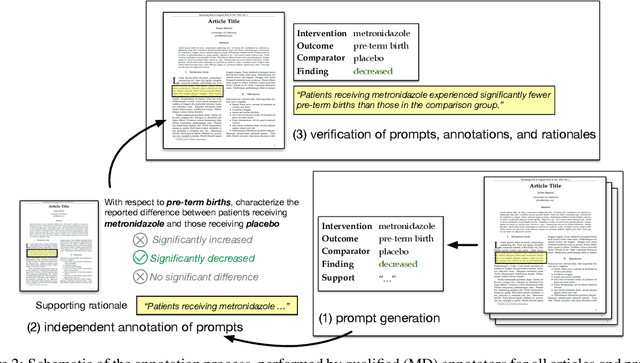
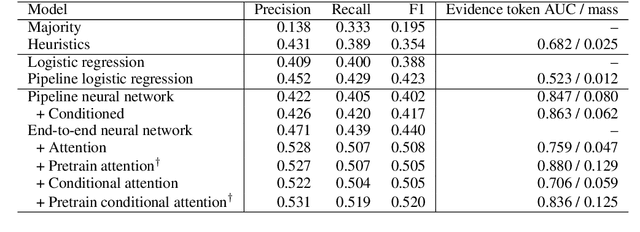
Abstract:How do we know if a particular medical treatment actually works? Ideally one would consult all available evidence from relevant clinical trials. Unfortunately, such results are primarily disseminated in natural language scientific articles, imposing substantial burden on those trying to make sense of them. In this paper, we present a new task and corpus for making this unstructured evidence actionable. The task entails inferring reported findings from a full-text article describing a randomized controlled trial (RCT) with respect to a given intervention, comparator, and outcome of interest, e.g., inferring if an article provides evidence supporting the use of aspirin to reduce risk of stroke, as compared to placebo. We present a new corpus for this task comprising 10,000+ prompts coupled with full-text articles describing RCTs. Results using a suite of models --- ranging from heuristic (rule-based) approaches to attentive neural architectures --- demonstrate the difficulty of the task, which we believe largely owes to the lengthy, technical input texts. To facilitate further work on this important, challenging problem we make the corpus, documentation, a website and leaderboard, and code for baselines and evaluation available at http://evidence-inference.ebm-nlp.com/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge