Sven Koitka
SALT: Introducing a Framework for Hierarchical Segmentations in Medical Imaging using Softmax for Arbitrary Label Trees
Jul 11, 2024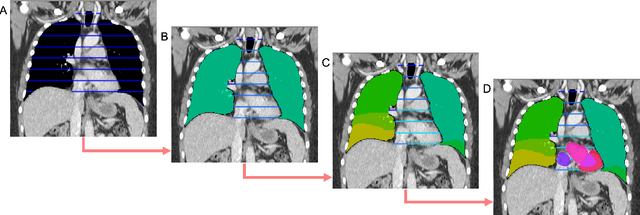
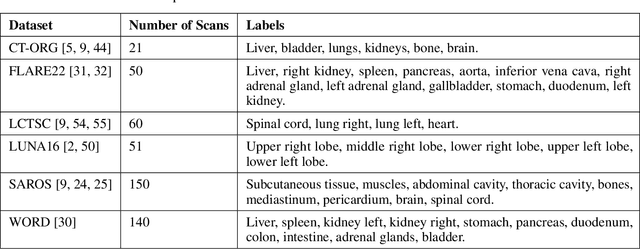
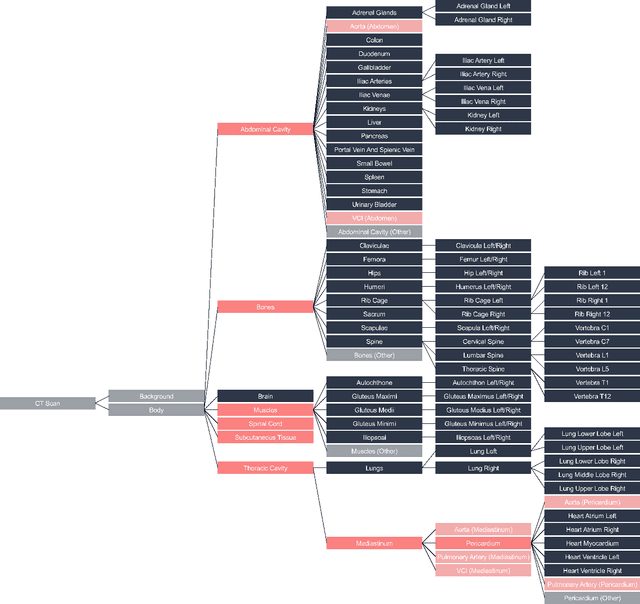
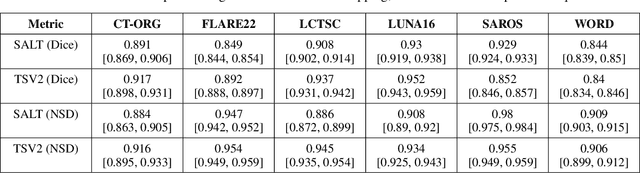
Abstract:Traditional segmentation networks approach anatomical structures as standalone elements, overlooking the intrinsic hierarchical connections among them. This study introduces Softmax for Arbitrary Label Trees (SALT), a novel approach designed to leverage the hierarchical relationships between labels, improving the efficiency and interpretability of the segmentations. This study introduces a novel segmentation technique for CT imaging, which leverages conditional probabilities to map the hierarchical structure of anatomical landmarks, such as the spine's division into lumbar, thoracic, and cervical regions and further into individual vertebrae. The model was developed using the SAROS dataset from The Cancer Imaging Archive (TCIA), comprising 900 body region segmentations from 883 patients. The dataset was further enhanced by generating additional segmentations with the TotalSegmentator, for a total of 113 labels. The model was trained on 600 scans, while validation and testing were conducted on 150 CT scans. Performance was assessed using the Dice score across various datasets, including SAROS, CT-ORG, FLARE22, LCTSC, LUNA16, and WORD. Among the evaluated datasets, SALT achieved its best results on the LUNA16 and SAROS datasets, with Dice scores of 0.93 and 0.929 respectively. The model demonstrated reliable accuracy across other datasets, scoring 0.891 on CT-ORG and 0.849 on FLARE22. The LCTSC dataset showed a score of 0.908 and the WORD dataset also showed good performance with a score of 0.844. SALT used the hierarchical structures inherent in the human body to achieve whole-body segmentations with an average of 35 seconds for 100 slices. This rapid processing underscores its potential for integration into clinical workflows, facilitating the automatic and efficient computation of full-body segmentations with each CT scan, thus enhancing diagnostic processes and patient care.
ROCOv2: Radiology Objects in COntext Version 2, an Updated Multimodal Image Dataset
May 16, 2024Abstract:Automated medical image analysis systems often require large amounts of training data with high quality labels, which are difficult and time consuming to generate. This paper introduces Radiology Object in COntext version 2 (ROCOv2), a multimodal dataset consisting of radiological images and associated medical concepts and captions extracted from the PMC Open Access subset. It is an updated version of the ROCO dataset published in 2018, and adds 35,705 new images added to PMC since 2018. It further provides manually curated concepts for imaging modalities with additional anatomical and directional concepts for X-rays. The dataset consists of 79,789 images and has been used, with minor modifications, in the concept detection and caption prediction tasks of ImageCLEFmedical Caption 2023. The dataset is suitable for training image annotation models based on image-caption pairs, or for multi-label image classification using Unified Medical Language System (UMLS) concepts provided with each image. In addition, it can serve for pre-training of medical domain models, and evaluation of deep learning models for multi-task learning.
Fully-automated Body Composition Analysis in Routine CT Imaging Using 3D Semantic Segmentation Convolutional Neural Networks
Feb 25, 2020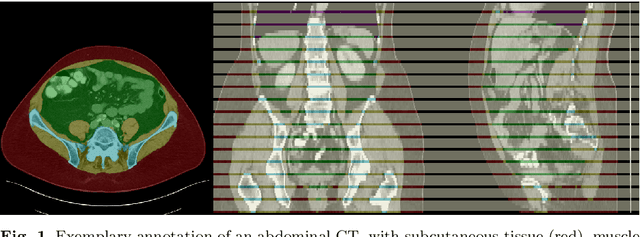
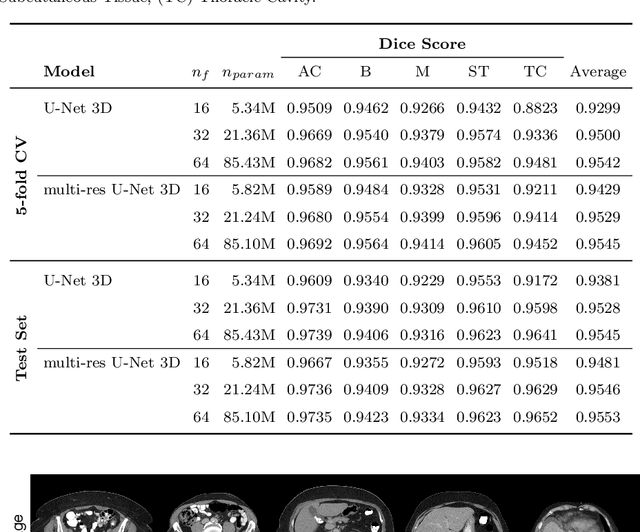
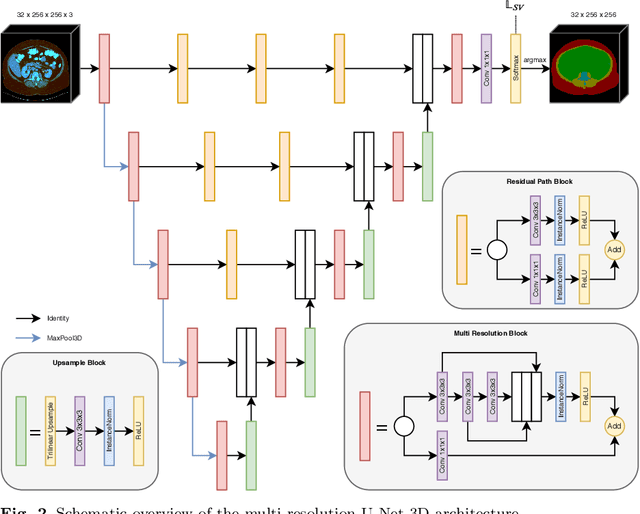
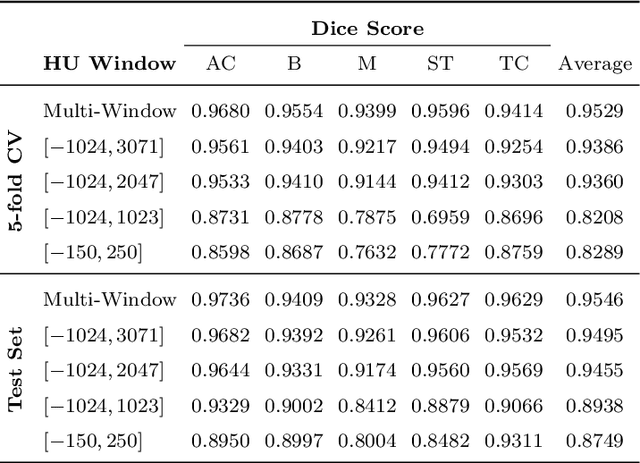
Abstract:Body tissue composition is a long-known biomarker with high diagnostic and prognostic value in cardiovascular, oncological and orthopaedic diseases, but also in rehabilitation medicine or drug dosage. In this study, the aim was to develop a fully automated, reproducible and quantitative 3D volumetry of body tissue composition from standard CT examinations of the abdomen in order to be able to offer such valuable biomarkers as part of routine clinical imaging. Therefore an in-house dataset of 40 CTs for training and 10 CTs for testing were fully annotated on every fifth axial slice with five different semantic body regions: abdominal cavity, bones, muscle, subcutaneous tissue, and thoracic cavity. Multi-resolution U-Net 3D neural networks were employed for segmenting these body regions, followed by subclassifying adipose tissue and muscle using known hounsfield unit limits. The S{\o}rensen Dice scores averaged over all semantic regions was 0.9553 and the intra-class correlation coefficients for subclassified tissues were above 0.99. Our results show that fully-automated body composition analysis on routine CT imaging can provide stable biomarkers across the whole abdomen and not just on L3 slices, which is historically the reference location for analysing body composition in the clinical routine.
Utilizing Neural Networks and Linguistic Metadata for Early Detection of Depression Indications in Text Sequences
Apr 19, 2018



Abstract:Depression is ranked as the largest contributor to global disability and is also a major reason for suicide. Still, many individuals suffering from forms of depression are not treated for various reasons. Previous studies have shown that depression also has an effect on language usage and that many depressed individuals use social media platforms or the internet in general to get information or discuss their problems. This paper addresses the early detection of depression using machine learning models based on messages on a social platform. In particular, a convolutional neural network based on different word embeddings is evaluated and compared to a classification based on user-level linguistic metadata. An ensemble of both approaches is shown to achieve state-of-the-art results in a current early detection task. Furthermore, the currently popular ERDE score as metric for early detection systems is examined in detail and its drawbacks in the context of shared tasks are illustrated. A slightly modified metric is proposed and compared to the original score. Finally, a new word embedding was trained on a large corpus of the same domain as the described task and is evaluated as well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge