Alberto Santamaria-Pang
Health AI, Microsoft, Redmond Washington
From Embeddings to Accuracy: Comparing Foundation Models for Radiographic Classification
May 16, 2025Abstract:Foundation models, pretrained on extensive datasets, have significantly advanced machine learning by providing robust and transferable embeddings applicable to various domains, including medical imaging diagnostics. This study evaluates the utility of embeddings derived from both general-purpose and medical domain-specific foundation models for training lightweight adapter models in multi-class radiography classification, focusing specifically on tube placement assessment. A dataset comprising 8842 radiographs classified into seven distinct categories was employed to extract embeddings using six foundation models: DenseNet121, BiomedCLIP, Med-Flamingo, MedImageInsight, Rad-DINO, and CXR-Foundation. Adapter models were subsequently trained using classical machine learning algorithms. Among these combinations, MedImageInsight embeddings paired with an support vector machine adapter yielded the highest mean area under the curve (mAUC) at 93.8%, followed closely by Rad-DINO (91.1%) and CXR-Foundation (89.0%). In comparison, BiomedCLIP and DenseNet121 exhibited moderate performance with mAUC scores of 83.0% and 81.8%, respectively, whereas Med-Flamingo delivered the lowest performance at 75.1%. Notably, most adapter models demonstrated computational efficiency, achieving training within one minute and inference within seconds on CPU, underscoring their practicality for clinical applications. Furthermore, fairness analyses on adapters trained on MedImageInsight-derived embeddings indicated minimal disparities, with gender differences in performance within 2% and standard deviations across age groups not exceeding 3%. These findings confirm that foundation model embeddings-especially those from MedImageInsight-facilitate accurate, computationally efficient, and equitable diagnostic classification using lightweight adapters for radiographic image analysis.
WaveFormer: A 3D Transformer with Wavelet-Driven Feature Representation for Efficient Medical Image Segmentation
Apr 01, 2025Abstract:Transformer-based architectures have advanced medical image analysis by effectively modeling long-range dependencies, yet they often struggle in 3D settings due to substantial memory overhead and insufficient capture of fine-grained local features. We address these limitations with WaveFormer, a novel 3D-transformer that: i) leverages the fundamental frequency-domain properties of features for contextual representation, and ii) is inspired by the top-down mechanism of the human visual recognition system, making it a biologically motivated architecture. By employing discrete wavelet transformations (DWT) at multiple scales, WaveFormer preserves both global context and high-frequency details while replacing heavy upsampling layers with efficient wavelet-based summarization and reconstruction. This significantly reduces the number of parameters, which is critical for real-world deployment where computational resources and training times are constrained. Furthermore, the model is generic and easily adaptable to diverse applications. Evaluations on BraTS2023, FLARE2021, and KiTS2023 demonstrate performance on par with state-of-the-art methods while offering substantially lower computational complexity.
Multi-Modal Mamba Modeling for Survival Prediction (M4Survive): Adapting Joint Foundation Model Representations
Mar 13, 2025Abstract:Accurate survival prediction in oncology requires integrating diverse imaging modalities to capture the complex interplay of tumor biology. Traditional single-modality approaches often fail to leverage the complementary insights provided by radiological and pathological assessments. In this work, we introduce M4Survive (Multi-Modal Mamba Modeling for Survival Prediction), a novel framework that learns joint foundation model representations using efficient adapter networks. Our approach dynamically fuses heterogeneous embeddings from a foundation model repository (e.g., MedImageInsight, BiomedCLIP, Prov-GigaPath, UNI2-h), creating a correlated latent space optimized for survival risk estimation. By leveraging Mamba-based adapters, M4Survive enables efficient multi-modal learning while preserving computational efficiency. Experimental evaluations on benchmark datasets demonstrate that our approach outperforms both unimodal and traditional static multi-modal baselines in survival prediction accuracy. This work underscores the potential of foundation model-driven multi-modal fusion in advancing precision oncology and predictive analytics.
MedImageInsight: An Open-Source Embedding Model for General Domain Medical Imaging
Oct 09, 2024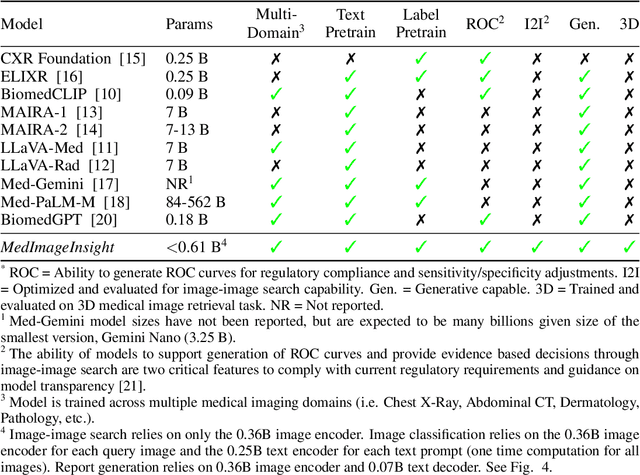
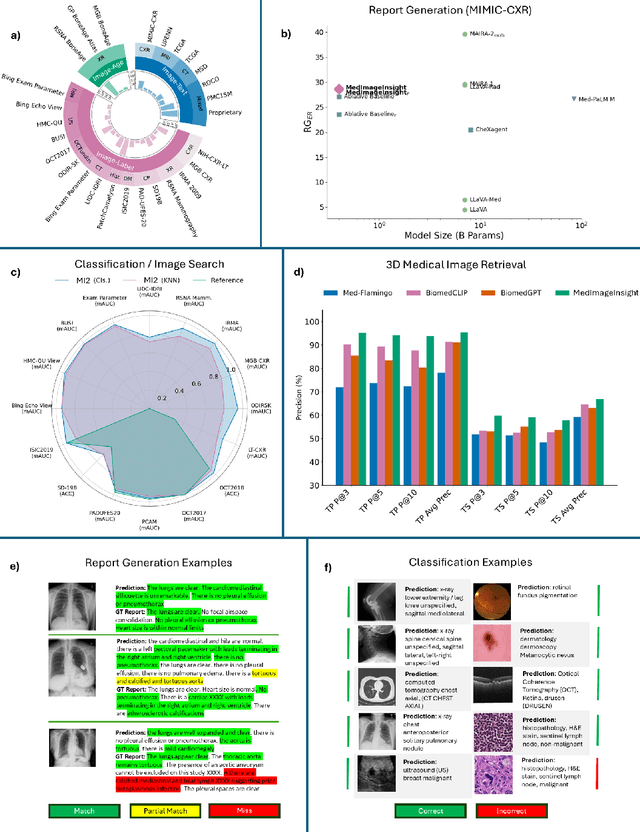
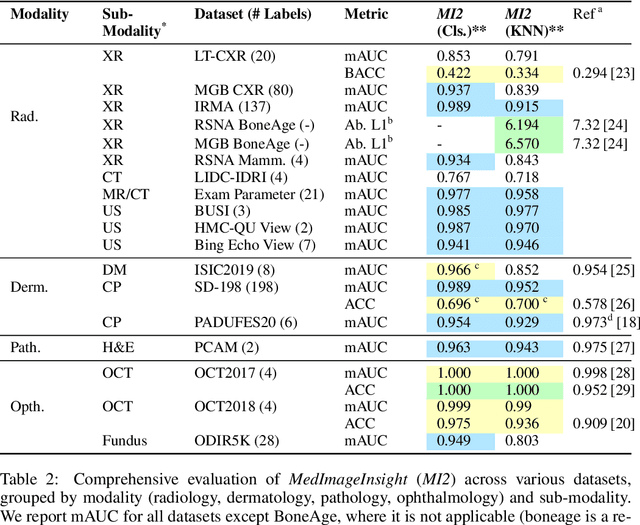
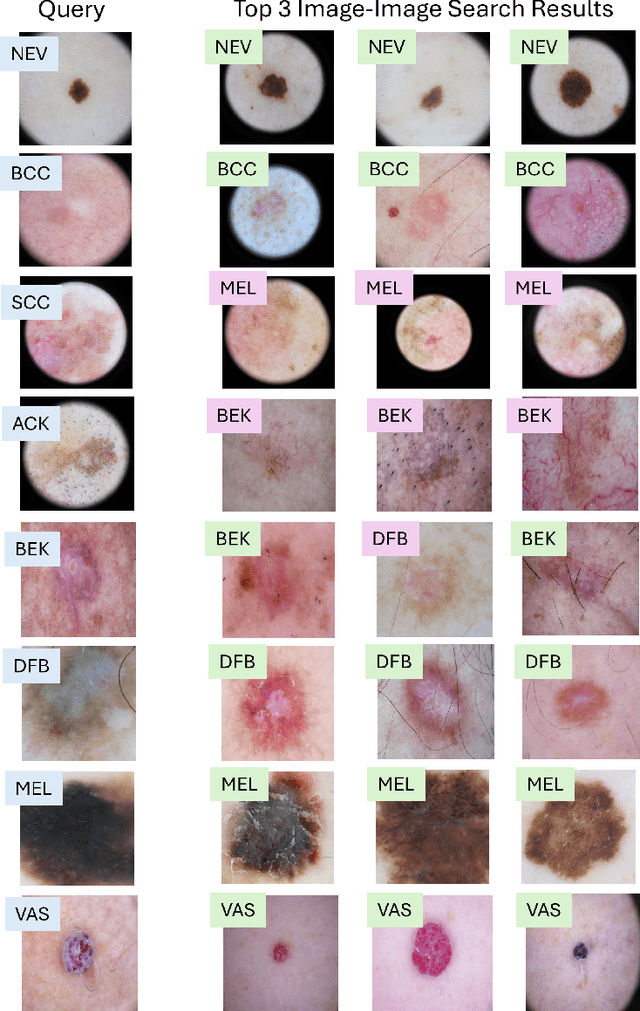
Abstract:In this work, we present MedImageInsight, an open-source medical imaging embedding model. MedImageInsight is trained on medical images with associated text and labels across a diverse collection of domains, including X-Ray, CT, MRI, dermoscopy, OCT, fundus photography, ultrasound, histopathology, and mammography. Rigorous evaluations demonstrate MedImageInsight's ability to achieve state-of-the-art (SOTA) or human expert level performance across classification, image-image search, and fine-tuning tasks. Specifically, on public datasets, MedImageInsight achieves SOTA in CT 3D medical image retrieval, as well as SOTA in disease classification and search for chest X-ray, dermatology, and OCT imaging. Furthermore, MedImageInsight achieves human expert performance in bone age estimation (on both public and partner data), as well as AUC above 0.9 in most other domains. When paired with a text decoder, MedImageInsight achieves near SOTA level single image report findings generation with less than 10\% the parameters of other models. Compared to fine-tuning GPT-4o with only MIMIC-CXR data for the same task, MedImageInsight outperforms in clinical metrics, but underperforms on lexical metrics where GPT-4o sets a new SOTA. Importantly for regulatory purposes, MedImageInsight can generate ROC curves, adjust sensitivity and specificity based on clinical need, and provide evidence-based decision support through image-image search (which can also enable retrieval augmented generation). In an independent clinical evaluation of image-image search in chest X-ray, MedImageInsight outperformed every other publicly available foundation model evaluated by large margins (over 6 points AUC), and significantly outperformed other models in terms of AI fairness (across age and gender). We hope releasing MedImageInsight will help enhance collective progress in medical imaging AI research and development.
Emergent Language Symbolic Autoencoder (ELSA) with Weak Supervision to Model Hierarchical Brain Networks
Apr 15, 2024Abstract:Brain networks display a hierarchical organization, a complexity that poses a challenge for existing deep learning models, often structured as flat classifiers, leading to difficulties in interpretability and the 'black box' issue. To bridge this gap, we propose a novel architecture: a symbolic autoencoder informed by weak supervision and an Emergent Language (EL) framework. This model moves beyond traditional flat classifiers by producing hierarchical clusters and corresponding imagery, subsequently represented through symbolic sentences to improve the clinical interpretability of hierarchically organized data such as intrinsic brain networks, which can be characterized using resting-state fMRI images. Our innovation includes a generalized hierarchical loss function designed to ensure that both sentences and images accurately reflect the hierarchical structure of functional brain networks. This enables us to model functional brain networks from a broader perspective down to more granular details. Furthermore, we introduce a quantitative method to assess the hierarchical consistency of these symbolic representations. Our qualitative analyses show that our model successfully generates hierarchically organized, clinically interpretable images, a finding supported by our quantitative evaluations. We find that our best performing loss function leads to a hierarchical consistency of over 97% when identifying images corresponding to brain networks. This approach not only advances the interpretability of deep learning models in neuroimaging analysis but also represents a significant step towards modeling the intricate hierarchical nature of brain networks.
Foundation Models for Biomedical Image Segmentation: A Survey
Jan 15, 2024Abstract:Recent advancements in biomedical image analysis have been significantly driven by the Segment Anything Model (SAM). This transformative technology, originally developed for general-purpose computer vision, has found rapid application in medical image processing. Within the last year, marked by over 100 publications, SAM has demonstrated its prowess in zero-shot learning adaptations for medical imaging. The fundamental premise of SAM lies in its capability to segment or identify objects in images without prior knowledge of the object type or imaging modality. This approach aligns well with tasks achievable by the human visual system, though its application in non-biological vision contexts remains more theoretically challenging. A notable feature of SAM is its ability to adjust segmentation according to a specified resolution scale or area of interest, akin to semantic priming. This adaptability has spurred a wave of creativity and innovation in applying SAM to medical imaging. Our review focuses on the period from April 1, 2023, to September 30, 2023, a critical first six months post-initial publication. We examine the adaptations and integrations of SAM necessary to address longstanding clinical challenges, particularly in the context of 33 open datasets covered in our analysis. While SAM approaches or achieves state-of-the-art performance in numerous applications, it falls short in certain areas, such as segmentation of the carotid artery, adrenal glands, optic nerve, and mandible bone. Our survey delves into the innovative techniques where SAM's foundational approach excels and explores the core concepts in translating and applying these models effectively in diverse medical imaging scenarios.
3D-MIR: A Benchmark and Empirical Study on 3D Medical Image Retrieval in Radiology
Nov 23, 2023



Abstract:The increasing use of medical imaging in healthcare settings presents a significant challenge due to the increasing workload for radiologists, yet it also offers opportunity for enhancing healthcare outcomes if effectively leveraged. 3D image retrieval holds potential to reduce radiologist workloads by enabling clinicians to efficiently search through diagnostically similar or otherwise relevant cases, resulting in faster and more precise diagnoses. However, the field of 3D medical image retrieval is still emerging, lacking established evaluation benchmarks, comprehensive datasets, and thorough studies. This paper attempts to bridge this gap by introducing a novel benchmark for 3D Medical Image Retrieval (3D-MIR) that encompasses four different anatomies imaged with computed tomography. Using this benchmark, we explore a diverse set of search strategies that use aggregated 2D slices, 3D volumes, and multi-modal embeddings from popular multi-modal foundation models as queries. Quantitative and qualitative assessments of each approach are provided alongside an in-depth discussion that offers insight for future research. To promote the advancement of this field, our benchmark, dataset, and code are made publicly available.
Region-based Contrastive Pretraining for Medical Image Retrieval with Anatomic Query
May 09, 2023



Abstract:We introduce a novel Region-based contrastive pretraining for Medical Image Retrieval (RegionMIR) that demonstrates the feasibility of medical image retrieval with similar anatomical regions. RegionMIR addresses two major challenges for medical image retrieval i) standardization of clinically relevant searching criteria (e.g., anatomical, pathology-based), and ii) localization of anatomical area of interests that are semantically meaningful. In this work, we propose an ROI image retrieval image network that retrieves images with similar anatomy by extracting anatomical features (via bounding boxes) and evaluate similarity between pairwise anatomy-categorized features between the query and the database of images using contrastive learning. ROI queries are encoded using a contrastive-pretrained encoder that was fine-tuned for anatomy classification, which generates an anatomical-specific latent space for region-correlated image retrieval. During retrieval, we compare the anatomically encoded query to find similar features within a feature database generated from training samples, and retrieve images with similar regions from training samples. We evaluate our approach on both anatomy classification and image retrieval tasks using the Chest ImaGenome Dataset. Our proposed strategy yields an improvement over state-of-the-art pretraining and co-training strategies, from 92.24 to 94.12 (2.03%) classification accuracy in anatomies. We qualitatively evaluate the image retrieval performance demonstrating generalizability across multiple anatomies with different morphology.
Deep Labeling of fMRI Brain Networks
May 05, 2023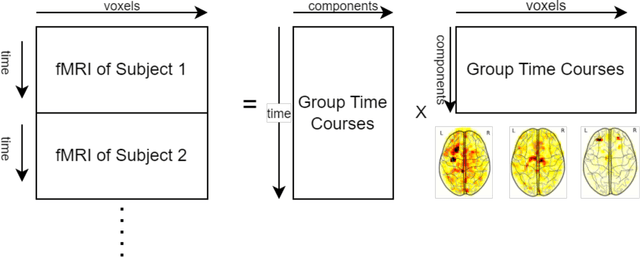
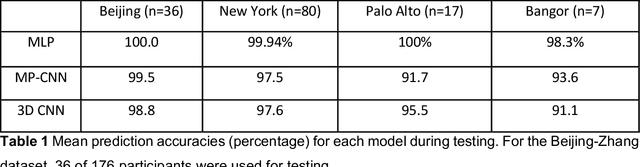
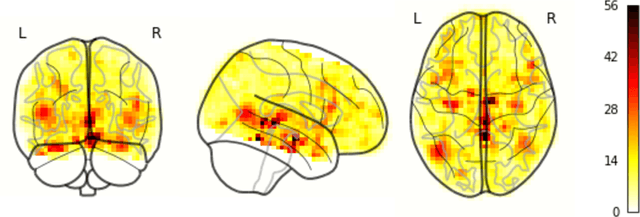
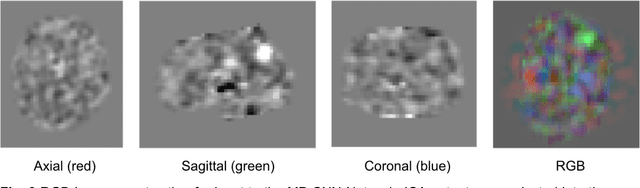
Abstract:Resting State Networks (RSNs) of the brain extracted from Resting State functional Magnetic Resonance Imaging (RS-fMRI) are used in the pre-surgical planning to guide the neurosurgeon. This is difficult, though, as expert knowledge is required to label each of the RSNs. There is a lack of efficient and standardized methods to be used in clinical workflows. Additionally, these methods need to be generalizable since the method needs to work well regardless of the acquisition technique. We propose an accurate, fast, and lightweight deep learning approach to label RSNs. Group Independent Component Analysis (ICA) was used to extract large scale functional connectivity patterns in the cohort and dual regression was used to back project them on individual subject RSNs. We compare a Multi-Layer Perceptron (MLP) based method with 2D and 3D Convolutional Neural Networks (CNNs) and find that the MLP is faster and more accurate. The MLP method performs as good or better than other works despite its compact size. We prove the generalizability of our method by showing that the MLP performs at 100% accuracy in the holdout dataset and 98.3% accuracy in three other sites' fMRI acquisitions.
Deep Labeling of fMRI Brain Networks Using Cloud Based Processing
Sep 20, 2022



Abstract:Resting state fMRI is an imaging modality which reveals brain activity localization through signal changes, in what is known as Resting State Networks (RSNs). This technique is gaining popularity in neurosurgical pre-planning to visualize the functional regions and assess regional activity. Labeling of rs-fMRI networks require subject-matter expertise and is time consuming, creating a need for an automated classification algorithm. While the impact of AI in medical diagnosis has shown great progress; deploying and maintaining these in a clinical setting is an unmet need. We propose an end-to-end reproducible pipeline which incorporates image processing of rs-fMRI in a cloud-based workflow while using deep learning to automate the classification of RSNs. We have architected a reproducible Azure Machine Learning cloud-based medical imaging concept pipeline for fMRI analysis integrating the popular FMRIB Software Library (FSL) toolkit. To demonstrate a clinical application using a large dataset, we compare three neural network architectures for classification of deeper RSNs derived from processed rs-fMRI. The three algorithms are: an MLP, a 2D projection-based CNN, and a fully 3D CNN classification networks. Each of the net-works was trained on the rs-fMRI back-projected independent components giving >98% accuracy for each classification method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge