Haris I Sair
Department of Radiology and Radiological Science, Johns Hopkins School of Medicine, Malone Center for Engineering in Healthcare, Johns Hopkins University
Emergent Language Symbolic Autoencoder (ELSA) with Weak Supervision to Model Hierarchical Brain Networks
Apr 15, 2024Abstract:Brain networks display a hierarchical organization, a complexity that poses a challenge for existing deep learning models, often structured as flat classifiers, leading to difficulties in interpretability and the 'black box' issue. To bridge this gap, we propose a novel architecture: a symbolic autoencoder informed by weak supervision and an Emergent Language (EL) framework. This model moves beyond traditional flat classifiers by producing hierarchical clusters and corresponding imagery, subsequently represented through symbolic sentences to improve the clinical interpretability of hierarchically organized data such as intrinsic brain networks, which can be characterized using resting-state fMRI images. Our innovation includes a generalized hierarchical loss function designed to ensure that both sentences and images accurately reflect the hierarchical structure of functional brain networks. This enables us to model functional brain networks from a broader perspective down to more granular details. Furthermore, we introduce a quantitative method to assess the hierarchical consistency of these symbolic representations. Our qualitative analyses show that our model successfully generates hierarchically organized, clinically interpretable images, a finding supported by our quantitative evaluations. We find that our best performing loss function leads to a hierarchical consistency of over 97% when identifying images corresponding to brain networks. This approach not only advances the interpretability of deep learning models in neuroimaging analysis but also represents a significant step towards modeling the intricate hierarchical nature of brain networks.
Applications of Sequential Learning for Medical Image Classification
Sep 26, 2023



Abstract:Purpose: The aim of this work is to develop a neural network training framework for continual training of small amounts of medical imaging data and create heuristics to assess training in the absence of a hold-out validation or test set. Materials and Methods: We formulated a retrospective sequential learning approach that would train and consistently update a model on mini-batches of medical images over time. We address problems that impede sequential learning such as overfitting, catastrophic forgetting, and concept drift through PyTorch convolutional neural networks (CNN) and publicly available Medical MNIST and NIH Chest X-Ray imaging datasets. We begin by comparing two methods for a sequentially trained CNN with and without base pre-training. We then transition to two methods of unique training and validation data recruitment to estimate full information extraction without overfitting. Lastly, we consider an example of real-life data that shows how our approach would see mainstream research implementation. Results: For the first experiment, both approaches successfully reach a ~95% accuracy threshold, although the short pre-training step enables sequential accuracy to plateau in fewer steps. The second experiment comparing two methods showed better performance with the second method which crosses the ~90% accuracy threshold much sooner. The final experiment showed a slight advantage with a pre-training step that allows the CNN to cross ~60% threshold much sooner than without pre-training. Conclusion: We have displayed sequential learning as a serviceable multi-classification technique statistically comparable to traditional CNNs that can acquire data in small increments feasible for clinically realistic scenarios.
Deep Labeling of fMRI Brain Networks
May 05, 2023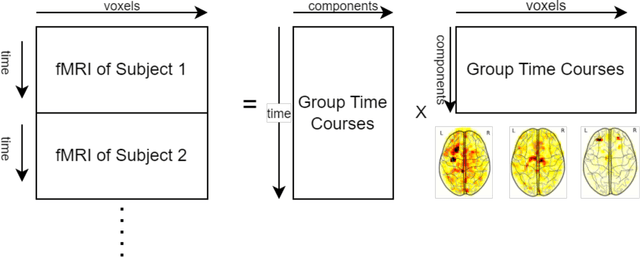
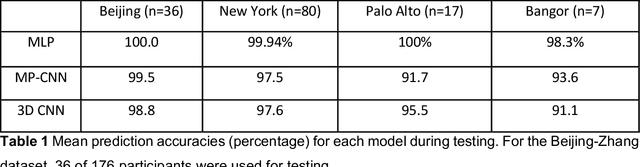
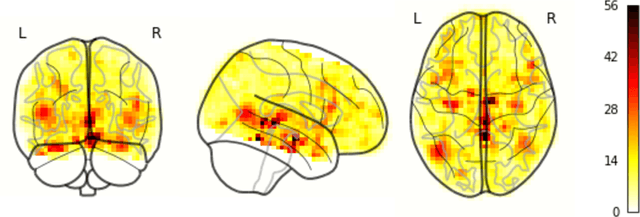
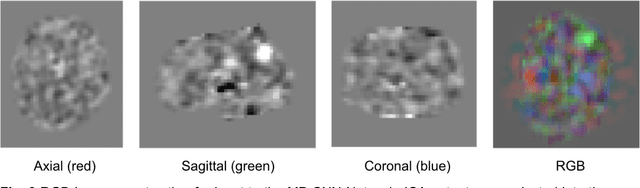
Abstract:Resting State Networks (RSNs) of the brain extracted from Resting State functional Magnetic Resonance Imaging (RS-fMRI) are used in the pre-surgical planning to guide the neurosurgeon. This is difficult, though, as expert knowledge is required to label each of the RSNs. There is a lack of efficient and standardized methods to be used in clinical workflows. Additionally, these methods need to be generalizable since the method needs to work well regardless of the acquisition technique. We propose an accurate, fast, and lightweight deep learning approach to label RSNs. Group Independent Component Analysis (ICA) was used to extract large scale functional connectivity patterns in the cohort and dual regression was used to back project them on individual subject RSNs. We compare a Multi-Layer Perceptron (MLP) based method with 2D and 3D Convolutional Neural Networks (CNNs) and find that the MLP is faster and more accurate. The MLP method performs as good or better than other works despite its compact size. We prove the generalizability of our method by showing that the MLP performs at 100% accuracy in the holdout dataset and 98.3% accuracy in three other sites' fMRI acquisitions.
Deep Labeling of fMRI Brain Networks Using Cloud Based Processing
Sep 20, 2022



Abstract:Resting state fMRI is an imaging modality which reveals brain activity localization through signal changes, in what is known as Resting State Networks (RSNs). This technique is gaining popularity in neurosurgical pre-planning to visualize the functional regions and assess regional activity. Labeling of rs-fMRI networks require subject-matter expertise and is time consuming, creating a need for an automated classification algorithm. While the impact of AI in medical diagnosis has shown great progress; deploying and maintaining these in a clinical setting is an unmet need. We propose an end-to-end reproducible pipeline which incorporates image processing of rs-fMRI in a cloud-based workflow while using deep learning to automate the classification of RSNs. We have architected a reproducible Azure Machine Learning cloud-based medical imaging concept pipeline for fMRI analysis integrating the popular FMRIB Software Library (FSL) toolkit. To demonstrate a clinical application using a large dataset, we compare three neural network architectures for classification of deeper RSNs derived from processed rs-fMRI. The three algorithms are: an MLP, a 2D projection-based CNN, and a fully 3D CNN classification networks. Each of the net-works was trained on the rs-fMRI back-projected independent components giving >98% accuracy for each classification method.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge